Page 65 - Clinical Immunology_ Principles and Practice ( PDFDrive )
P. 65
50 ParT ONE Principles of Immune Response
Bacteria
MDP
RIP2
TAK1 IKK LRRs
CARD9 NOD2
CARD CARD NOD
IκB
MAPKs NF-κB IRF7
IRF3
Viral RNA
Nucleus TBK1/IKKι
IL-1/18
IκB
RIG-1
NF-κB IPS-1
Pro-IL-1/18
MDA5
CARD Caspase1 Mitochondria
CARD
ASC
PYD
PAMPs
PYD NOD LRR NLRP3 DAMPs
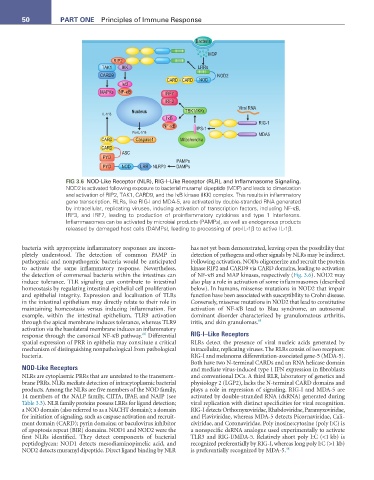
FIG 3.6 NOD-Like Receptor (NLR), RIG-I–Like Receptor (RLR), and Inflammasome Signaling.
NOD2 is activated following exposure to bacterial muramyl dipeptide (MDP) and leads to dimerization
and activation of RIP2, TAK1, CARD9, and the IκB kinase (IKK) complex. This results in inflammatory
gene transcription. RLRs, like RIG-I and MDA-5, are activated by double-stranded RNA generated
by intracellular, replicating viruses, inducing activation of transcription factors, including NF-κB,
IRF3, and IRF7, leading to production of proinflammatory cytokines and type 1 interferons.
Inflammasomes can be activated by microbial products (PAMPs), as well as endogenous products
released by damaged host cells (DAMPs), leading to processing of pro-IL-1β to active IL-1β.
bacteria with appropriate inflammatory responses are incom- has not yet been demonstrated, leaving open the possibility that
pletely understood. The detection of common PAMP in detection of pathogens and other signals by NLRs may be indirect.
pathogenic and nonpathogenic bacteria would be anticipated Following activation, NODs oligomerize and recruit the protein
to activate the same inflammatory response. Nevertheless, kinase RIP2 and CARD9 via CARD domains, leading to activation
the detection of commensal bacteria within the intestines can of NF-κB and MAP kinases, respectively (Fig. 3.6). NOD2 may
induce tolerance. TLR signaling can contribute to intestinal also play a role in activation of some inflammasomes (described
homeostasis by regulating intestinal epithelial cell proliferation below). In humans, missense mutations in NOD2 that impair
and epithelial integrity. Expression and localization of TLRs function have been associated with susceptibility to Crohn disease.
in the intestinal epithelium may directly relate to their role in Conversely, missense mutations in NOD2 that lead to constitutive
maintaining homeostasis versus inducing inflammation. For activation of NF-κB lead to Blau syndrome, an autosomal
example, within the intestinal epithelium, TLR9 activation dominant disorder characterized by granulomatous arthritis,
through the apical membrane induces tolerance, whereas TLR9 iritis, and skin granulomas. 61
activation via the basolateral membrane induces an inflammatory
60
response through the canonical NF-κB pathway. Differential RIG-I–Like Receptors
spatial expression of PRR in epithelia may constitute a critical RLRs detect the presence of viral nucleic acids generated by
mechanism of distinguishing nonpathological from pathological intracellular, replicating viruses. The RLRs consist of two receptors:
bacteria. RIG-I and melanoma differentiation-associated gene-5 (MDA-5).
Both have two N-terminal CARDs and an RNA helicase domain
NOD-Like Receptors and mediate virus-induced type 1 IFN expression in fibroblasts
NLRs are cytoplasmic PRRs that are unrelated to the transmem- and conventional DCs. A third RLR, laboratory of genetics and
brane PRRs. NLRs mediate detection of intracytoplasmic bacterial physiology 2 (LGP2), lacks the N-terminal CARD domains and
products. Among the NLRs are five members of the NOD family, plays a role in repression of signaling. RIG-I and MDA-5 are
14 members of the NALP family, CIITA, IPAF, and NAIP (see activated by double-stranded RNA (dsRNA) generated during
Table 3.3). NLR family proteins possess LRRs for ligand detection; viral replication with distinct specificities for viral recognition.
a NOD domain (also referred to as a NACHT domain); a domain RIG-I detects Orthomyxoviridae, Rhabdoviridae, Paramyxoviridae,
for initiation of signaling, such as caspase activation and recruit- and Flaviviridae, whereas MDA-5 detects Picornaviridae, Cali-
ment domain (CARD); pyrin domains; or baculovirus inhibitor civiridae, and Coronaviridae. Poly inosine:cytosine (poly I:C) is
of apoptosis repeat (BIR) domains. NOD1 and NOD2 were the a nonspecific dsRNA analogue used experimentally to activate
first NLRs identified. They detect components of bacterial TLR3 and RIG-I/MDA-5. Relatively short poly I:C (<1 kb) is
peptidoglycan: NOD1 detects mesodiaminopimelic acid, and recognized preferentially by RIG-I, whereas long poly I:C (>1 kb)
NOD2 detects muramyl dipeptide. Direct ligand binding by NLR is preferentially recognized by MDA-5. 48