Page 97 - Clinical Immunology_ Principles and Practice ( PDFDrive )
P. 97
82 PARt oNE Principles of Immune Response
TABLE 5.1 Peptide-Binding Motifs
Encoded by Different HLA Alleles Influence
the Number of Peptides in a Protein
that can be Recognized by a HLA Molecule
α 1 (e.g., HIV Envelope Protein)
Allele designation HLA-B*27:05 HLA-B*35:01 HLA-B*07:02
Peptide-binding XRXXXXXX[KRYL] XPXXXXXXY XPXXXXXXL
motif
Peptides from the IRGKVQKEY None DPNPQEVVL
C HIV envelope IRPVVSTQL KPCVKLTPL
N protein able to TRPNNNTRK RPVVSTQLL
bind to each IRIQRGPGR SPLSFQTHL
allotype SRAKWNNTL IPRRIRQGL
LREQFGNNK
FRPGGGDMR
WRSELYKYK
α 2
KRRVVQREK
ARILAVERY
N
ERDRDRSIR
LRSLCLFSY
TRIVELLGR
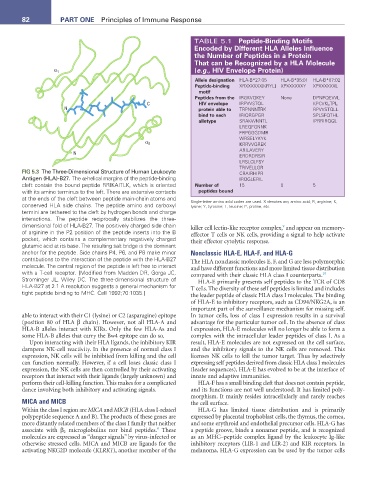
FIG 5.3 The Three-Dimensional Structure of Human Leukocyte CRAIRHIPR
Antigen (HLA)-B27. The α-helical margins of the peptide-binding IRQGLERIL
cleft contain the bound peptide RRIKAITLK, which is oriented Number of 15 0 5
with its amino terminus to the left. There are extensive contacts peptides bound
at the ends of the cleft between peptide main-chain atoms and
conserved HLA side chains. The peptide amino and carboxyl Single-letter amino acid codes are used. X denotes any amino acid; R, arginine; K,
lysine; Y, tyrosine; L, leucine; P, proline, etc.
termini are tethered to the cleft by hydrogen bonds and charge
interactions. The peptide reciprocally stabilizes the three-
dimensional fold of HLA-B27. The positively charged side chain killer cell lectin-like receptor complex, and appear on memory-
9
of arginine in the P2 position of the peptide inserts into the B effector T cells or NK cells, providing a signal to help activate
pocket, which contains a complementary negatively charged their effector cytolytic response.
glutamic acid at its base. The resulting salt bridge is the dominant
anchor for the peptide. Side chains P4, P6, and P8 make minor Nonclassic HLA-E, HLA-F, and HLA-G
contributions to the interaction of the peptide with the HLA-B27 The HLA nonclassic molecules E, F, and G are less polymorphic
molecule. The central region of the peptide is left free to interact and have different functions and more limited tissue distribution
with a T-cell receptor. [Modified from Madden DR, Gorga JC, compared with their classic HLA class I counterparts. 10
Strominger JL, Wiley DC. The three-dimensional structure of HLA-E primarily presents self peptides to the TCR of CD8
HLA-B27 at 2.1 A resolution suggests a general mechanism for T cells. The diversity of these self peptides is limited and includes
tight peptide binding to MHC. Cell 1992;70:1035.] the leader peptide of classic HLA class I molecules. The binding
of HLA-E to inhibitory receptors, such as CD94/NKG2A, is an
important part of the surveillance mechanism for missing self.
able to interact with their C1 (lysine) or C2 (asparagine) epitope In tumor cells, loss of class I expression results in a survival
(position 80 of HLA β chain). However, not all HLA-A and advantage for the particular tumor cell. In the absence of class
HLA-B alleles interact with KIRs. Only the few HLA-As and I expression, HLA-E molecules will no longer be able to form a
some HLA-B alleles that carry the Bw4 epitope can do so. complex with the intracellular leader peptides of class I. As a
Upon interacting with their HLA ligands, the inhibitory KIR result, HLA-E molecules are not expressed on the cell surface,
dampens NK-cell reactivity. In the presence of normal class I and the inhibitory signals to the NK cells are removed. This
expression, NK cells will be inhibited from killing and the cell licenses NK cells to kill the tumor target. Thus by selectively
can function normally. However, if a cell loses classic class I expressing self peptides derived from classic HLA class I molecules
expression, the NK cells are then controlled by their activating (leader sequences), HLA-E has evolved to be at the interface of
receptors that interact with their ligands (largely unknown) and innate and adaptive immunities.
perform their cell-killing function. This makes for a complicated HLA-F has a small binding cleft that does not contain peptide,
dance involving both inhibitory and activating signals. and its functions are not well understood. It has limited poly-
morphism. It mainly resides intracellularly and rarely reaches
MICA and MICB the cell surface.
Within the class I region are MICA and MICB (HLA class I-related HLA-G has limited tissue distribution and is primarily
polypeptide sequence A and B). The products of these genes are expressed by placental trophoblast cells, the thymus, the cornea,
more distantly related members of the class I family that neither and some erythroid and endothelial precursor cells. HLA-G has
8
associate with β 2 microglobulins nor bind peptides. These a peptide groove, binds a nonamer peptide, and is recognized
molecules are expressed as “danger signals” by virus-infected or as an MHC–peptide complex ligand by the leukocyte Ig-like
otherwise stressed cells. MICA and MICB are ligands for the inhibitory receptors (LIR-1 and LIR-2) and KIR receptors. In
activating NKG2D molecule (KLRK1), another member of the melanoma, HLA-G expression can be used by the tumor cells