Page 915 - Hematology_ Basic Principles and Practice ( PDFDrive )
P. 915
798 Part VII Hematologic Malignancies
1 2 3 4 5
5' JAK2 (RP11-3H3)
3' JAK2 (RP11-28A9)
6 7 8 9 10 11 12
13 14 15 16 17 18
CDKN2A
Centromere 9
19 20 21 22 X Y
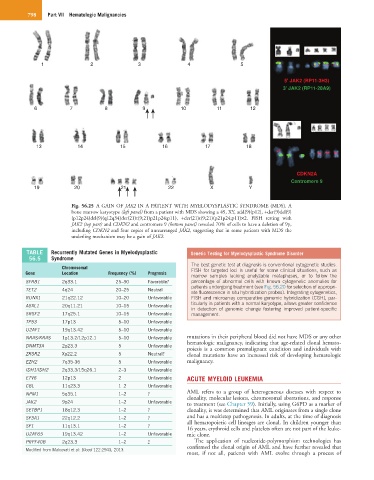
Fig. 56.25 A GAIN OF JAK2 IN A PATIENT WITH MYELODYSPLASTIC SYNDROME (MDS). A
bone marrow karyotype (left panel) from a patient with MDS showing a 49, XY, add(9)(p12), +der(9)del(9)
(p12p24)del(9)(q12q34)der(21)t(9;21)p21p24;p11), +der(21)t(9;21)(p21p24;p11)×2. FISH testing with
JAK2 (top part) and CDKN2 and centromere 9 (bottom panel) revealed 70% of cells to have a deletion of 9p,
including CDKN2 and four copies of unrearranged JAK2, suggesting that in some patients with MDS the
underling mechanism may be a gain of JAK2.
TABLE Recurrently Mutated Genes in Myelodysplastic Genetic Testing for Myelodysplastic Syndrome Disorder
56.5 Syndrome
The best genetic test at diagnosis is conventional cytogenetic studies.
Chromosomal FISH for targeted loci is useful for some clinical situations, such as
Gene Location Frequency (%) Prognosis
marrow samples lacking analyzable metaphases, or to follow the
SFRB1 2q33.1 25–30 Favorable? percentage of abnormal cells with known cytogenetic anomalies for
patients undergoing treatment (see Fig. 56.20 for selection of appropri-
TET2 4q24 20–25 Neutral
ate fluorescence in situ hybridization probes). Integrating cytogenetics,
RUNX1 21q22.12 10–20 Unfavorable FISH and microarray comparative genomic hybridization (CGH), par-
ASXL1 20q11.21 10–15 Unfavorable ticularly in patients with a normal karyotype, allows greater confidence
in detection of genomic change fostering improved patient-specific
SRSF2 17q25.1 10–15 Unfavorable management.
TP53 17p13 5–10 Unfavorable
U2AF1 19q13.42 5–10 Unfavorable
NRAS/KRAS 1p13.2/12p12.1 5–10 Unfavorable mutations in their peripheral blood did not have MDS or any other
DNMT3A 2p23.3 5 Unfavorable hematologic malignancy, indicating that age-related clonal hemato-
poiesis is a common premalignant condition and individuals with
ZRSR2 Xp22.2 5 Neutral? clonal mutations have an increased risk of developing hematologic
EZH2 7q35-36 5 Unfavorable malignancy.
IDH1/IDH2 2q33.3/15q26.1 2–3 Unfavorable
ETV6 12p13 2 Unfavorable ACUTE MYELOID LEUKEMIA
CBL 11q23.3 1–2 Unfavorable
NPM1 5q35.1 1–2 ? AML refers to a group of heterogeneous diseases with respect to
clonality, molecular lesions, chromosomal aberrations, and response
JAK2 9p24 1–2 Unfavorable to treatment (see Chapter 59). Initially, using G6PD as a marker of
SETBP1 18q12.3 1–2 ? clonality, it was determined that AML originates from a single clone
SF3A1 22q12.2 1–2 ? and has a multistep pathogenesis. In adults, at the time of diagnosis
all hematopoietic cell lineages are clonal. In children younger than
SF1 11q13.1 1–2 ?
16 years, erythroid cells and platelets often are not part of the leuke-
U2AF65 19q13.42 1–2 Unfavorable mic clone.
PRPF40B 2q23.3 1–2 ? The application of nucleotide-polymorphism technologies has
Modified from Malcovati et al: Blood 122:2943, 2013. confirmed the clonal origin of AML and have further revealed that
most, if not all, patients with AML evolve through a process of