Page 943 - Hematology_ Basic Principles and Practice ( PDFDrive )
P. 943
826 Part VII Hematologic Malignancies
21
hematopoietic cells before B-cell differentiation. These observations
may indicate that CLL develops from preleukemic multipotent
hematopoietic progenitors as a result of deregulation of B-cell recep- chr.5
tor intracellular signaling and that CLL develops from a preleukemic
phase. chr.7
chr.18
Genome Complexity in Chronic Lymphocytic Leukemia
chr.5
Most patients with CLL have two chromosomal abnormalities sug-
gesting genomic stability at the microscope level. Solid evidence exists
that chromosomal translocations detected by conventional cytogenet- chr.7
ics independently predict treatment failure, treatment-free survival,
and OS in untreated and treated patients with B-cell CLL. When chr.22
CLL-derived metaphase cells were obtained following systematic
stimulation using B-cell mitogens and activators, balanced and unbal- A B
anced translocations were observed in 34% to 42% of patients,
respectively. Following multivariate analysis, unbalanced transloca-
tions have independently been associated with risk for treatment
failure. Because del(13q) usually is cryptic by conventional cytogenet-
ics but easily detected with FISH, both conventional cytogenetics and
interphase FISH should be performed at baseline in patients with
advanced CLL.
Array CGH and NGS studies have effectively clarified the level
of genomic complexity in CLL and revealed that the average number
of mutations in CLL cells at diagnosis of CLL lies between 10 and
20, which is one of the lowest among the adult cancers, confirming
previous cytogenetic observations. The recent studies also revealed
that no single unifying mutation is responsible for CLL. Array-based
genomic profiling has extended previous cytogenetic data demon-
strating that a subset of patients with CLL have complex genomic
profiles pointing to the interplay between chromosomal abnormali-
ties and somatic mutations that are associated with reduced OS.
Moreover, telomere dysfunction in CLL and acute telomere attrition
results in fusion events that contribute to genomic complexity such
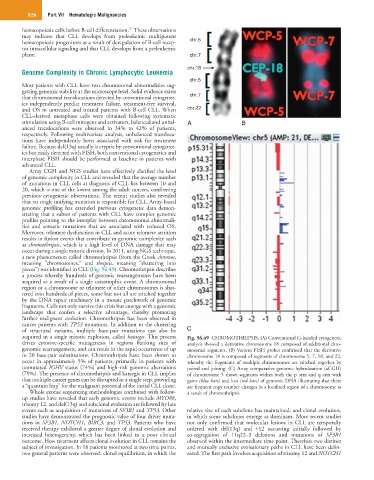
as chromothripsis, which is a high level of DNA damage that may
occur during a single mitotic division. In 2011, using NGS technique,
a new phenomenon called chromothripsis (from the Greek chromos,
meaning “chromosomes,” and thripsis, meaning “shattering into
pieces”) was identified in CLL (Fig. 56.49). Chromothripsis describes
a process whereby hundreds of genomic rearrangements have been
acquired as a result of a single catastrophic event. A chromosomal
region or a chromosome or telomere of other chromosomes is shat-
tered into hundreds of pieces, some but not all are stitched together
by the DNA repair machinery in a mosaic patchwork of genomic
fragments. Cells not only survive this crisis but emerge with a genomic
landscape that confers a selective advantage, thereby promoting
further malignant evolution. Chromothripsis has been observed in
cancer patients with TP53 mutations. In addition to the clustering
of structural variants, multiple base-pair mutations can also be C
acquired in a single mitotic explosion, called kataegis. This process Fig. 56.49 CHROMOTHRIPSIS. (A) Conventional G-banded cytogenetic
drives cytosine-specific mutagenesis in regions flanking sites of analysis showed a derivative chromosome 18 composed of additional chro-
genomic rearrangement, and can result in the rapid occurrence of up mosomal segments. (B) Various FISH probes confirmed that the derivative
to 20 base-pair substitutions. Chromothripsis have been shown to chromosome 18 is composed of segments of chromosome 5, 7, 18, and 22,
occur in approximately 5% of patients, primarily in patients with whereby the fragments of multiple chromosomes are stitched together by
unmutated IGHV status (74%) and high-risk genomic aberrations paired end joining. (C) Array comparative genomic hybridization (aCGH)
(79%). The presence of chromothripsis and kataegis in CLL implies of chromosome 5 shows segments within both the p arm and q arm with
that multiple cancer genes can be disrupted in a single step, providing gains (blue bars) and loss (red bars) of genomic DNA illustrating that there
a “quantum leap” for the malignant potential of the initial CLL clone. are frequent copy number changes in a localized region of a chromosome as
Whole exome sequencing methodologies combined with follow- a result of chromothripsis.
up studies have revealed that early genomic events include MYD88,
trisomy 12, and del(13q) and subclonal evolution are followed by late
events such as acquisition of mutations of SF3B1 and TP53. Other relative size of each subclone has maintained, and clonal evolution,
studies have demonstrated the prognostic value of four driver muta- in which some subclones emerge as dominant. More recent studies
tions in SF3B1, NOTCH1, BIRC3, and TP53. Patients who have not only confirmed that molecular lesions in CLL are temporally
received therapy exhibited a greater degree of clonal evolution and ordered with del(13q) and +12 occurring initially followed by
increased heterogeneity, which has been linked to a poor clinical co-segregation of 11q22–3 deletions and mutations of SF3B1
outcome. How treatment affects clonal evolution in CLL remains the observed within the intermediate time point. Therefore two distinct
subject of investigation. In 18 patients monitored at two time points, and mutually exclusive evolutionary paths in CLL have been delin-
two general patterns were observed: clonal equilibrium, in which the eated. The first path involves acquisition of trisomy 12 and NOTCH1