Page 961 - Hematology_ Basic Principles and Practice ( PDFDrive )
P. 961
844 Part VII Hematologic Malignancies
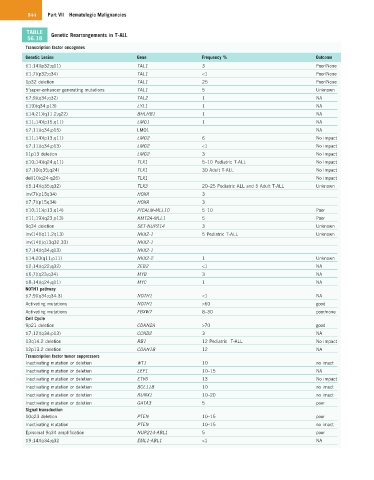
TABLE Genetic Rearrangements in T-ALL
56.18
Transcription factor oncogenes
Genetic Lesion Gene Frequency % Outcome
t(1;14)(p32;q11) TAL1 3 Poor/None
t(1;7)(p32;q34) TAL1 <1 Poor/None
1p32 deletion TAL1 25 Poor/None
5’super-enhancer generating mutations TAL1 5 Unknown
t(7;9)(q34;q32) TAL2 1 NA
t(19)(q34;p13) LYL1 1 NA
t(14;21)(q11.2;q22) BHLHB1 1 NA
t(11;14)(p15;q11) LMO1 1 NA
t(7;11)(q34;p15) LMO1 NA
t(11;14)(p13;q11) LMO2 6 No impact
t(7;11)(q34;p13) LMO2 <1 No impact
11p13 deletion LMO2 3 No impact
t(10;14)(q24;q11) TLX1 5–10 Pediatric T-ALL No impact
t(7;10(q35;q24) TLX1 30 Adult T-ALL No impact
del(10(q24-q26) TLX1 No impact
t(5;14)(q35;q32) TLX3 20–25 Pediatric ALL and 5 Adult T-ALL Unknown
inv(7)(p15q34) HOXA 3
t(7;7)(p15q34) HOXA 3
t(10;11)(p13;q14) PICALM-MLL10 5–10 Poor
t(11;19)(q23;p13) KMT2A-MLL1 5 Poor
9q34 deletion SET-NUP214 3 Unknown
inv(14)(q11.2q13) NKX2-1 5 Pediatric T-ALL Unknown
inv(14)((q13q32.33) NKX2-1
t(7;14)(q34;q13) NKX2-1
t(14;20(q11;p11) NKX2-2 1 Unknown
t(2;14)(q22;q32) ZEB2 <1 NA
t(6;7)(q23;q34) MYB 3 NA
t(8;14)(q24;q11) MYC 1 NA
NOTH1 pathway
t(7;9)(q34;q34.3) NOTH1 <1 NA
Activating mutations NOTH1 >60 good
Activating mutations FBXW7 8–30 poor/none
Cell Cycle
9p21 deletion CDKN2A >70 good
t(7;12)(q34;p13) CCND2 3 NA
13q14.2 deletion RB1 12 Pediatric T-ALL No impact
12p13.2 deletion CDKN1B 12 NA
Transcription factor tumor suppressors
Inactivating mutation or deletion WT1 10 no imact
Inactivating mutation or deletion LEF1 10–15 NA
Inactivating mutation or deletion ETV6 13 No impact
Inactivating mutation or deletion BCL11B 10 no imact
Inactivating mutation or deletion RUNX1 10–20 no imact
Inactivating mutation or deletion GATA3 5 poor
Signal transduction
10q23 deletion PTEN 10–15 poor
Inactivating mutation PTEN 10–15 no imact
Episomal 9q34 amplification NUP214-ABL1 5 poor
t(9;14)(q34;q32 EML1-ABL1 <1 NA