Page 963 - Hematology_ Basic Principles and Practice ( PDFDrive )
P. 963
846 Part VII Hematologic Malignancies
on the Y chromosome allows identification of male cells in the length polymorphism, STR analysis, or FISH XY analysis alone may
background of 100,000 female cells, providing a high sensitivity for not definitively assign the origin of the leukemic clone because
mixed chimerism. However, this approach is limited to the 50% of genomic deletions or amplifications of chromosomal segments may
patients who receive sex-mismatched transplants. Nevertheless, it occur during transplantation or disease process. The increased use of
remains the most sensitive and the fastest method of chimerism unrelated cord blood as a source of stem cells for allogeneic HCT
analysis, providing reliable quantitative results within 2 hours. raises the concern that hematopoietic progenitors containing preleu-
Detection of SNPs by chimerism analysis (SNP-PCR) is highly kemic clonal molecular rearrangements may be inadvertently trans-
sensitive. In one study using 11 different SNP loci, SNP-PCR analysis planted. Systematic screening of unselected cord blood samples
identified independent predictors of relapse after HCT. The two most revealed putative preleukemic rearrangements such as ETV6-RUNX1
commonly used methods for detection of chimerism after HCT are and RUNX1-RUNXT1. In a study of 1417 umbilical cord blood
fluorescence-based PCR amplification of short tandem repeats (STR- samples evidence for the ETV6 or RUNX1/RUNXT1 fusions were
PCR) and interphase FISH. Both methods are accurate and repro- not found. However, recent reports comparing clinical characteristics
ducible. The sensitivity of both methods approaches 1%; however, of donor-cell derived leukemia (DCL) from the standpoint of the
STR-PCR is sex independent and can be applied to all patients. FISH transplant source, with umbilical cord blood and bone marrow,
analysis, on the other hand, permits simultaneous evaluation of showed in some studies, that AML and MDS were recognized more
chimerism and residual disease in the same cell when high sensitivity frequently in DCL after cord blood transplant, but not in other
is not a requirement. FISH analysis for diagnostic genomic abnor- studies, whereas the incidence of AML and ALL was similar after
malities in conjunction with conventional cytogenetics remains bone marrow transplant. The median duration between the occur-
useful and reliable in determining the presence of residual disease rence of DCL following cord blood and bone marrow transplant was
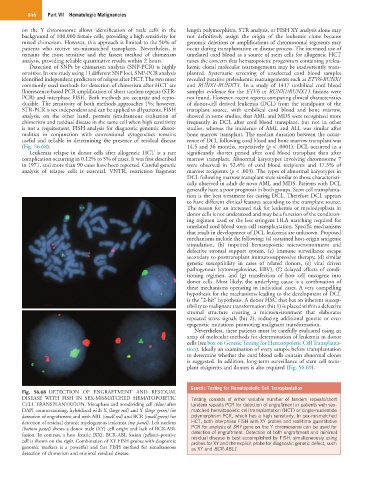
(Fig. 56.68). 14.5 and 36 months, respectively (p < .0001). DCL occurred in a
Leukemia relapse in donor cells after allogeneic HCT is a rare significantly shorter period after cord blood transplant than after
complication occurring in 0.12% to 5% of cases. It was first described marrow transplant. Abnormal karyotypes involving chromosome 7
in 1971, and more than 90 cases have been reported. Careful genetic were observed in 52.4% of cord blood recipients and 17.3% of
analysis of relapse cells is essential. VNTR, restriction fragment marrow recipients (p < .003). The types of abnormal karyotypes in
DCL following marrow transplant were similar to those characteristi-
cally observed in adult de novo AML and MDS. Patients with DCL
generally have a poor prognosis in both groups. Stem cell transplanta-
tion is the best treatment for curing DCL. Therefore DCL appears
to have different clinical features according to the transplant source.
The reason for an increased risk for leukemia or myelodysplasia in
donor cells is not understood and may be a function of the condition-
ing regimen used or the less stringent HLA matching required for
unrelated cord blood stem cell transplantation. Specific mechanisms
that result in development of DCL leukemia are unknown. Proposed
mechanisms include the following: (a) sustained host-origin antigenic
stimulation, (b) impaired hematopoietic microenvironment and
defective stromal support system, (c) immune surveillance escape
secondary to posttransplant immunosuppressive therapy, (d) similar
genetic susceptibility in cases of related donors, (e) viral driven
pathogenesis (cytomegalovirus, EBV), (f) delayed effects of condi-
tioning regimen, and (g) transfection of host cell oncogene into
donor cells. Most likely, the underlying cause is a combination of
these mechanisms operating in individual cases. A very compelling
hypothesis for the mechanisms leading to the development of DCL
is the “2-hit” hypothesis. A donor HSC that has an inherent suscep-
tibility to malignant transformation (hit 1) is placed within a defective
stromal structure creating a microenvironment that elaborates
repeated stress signals (hit 2), inducing additional genetic or even
epigenetic mutations promoting malignant transformation.
Nevertheless, these patients must be carefully evaluated using an
array of molecular methods for determination of leukemia in donor
cells (see box on Genetic Testing for Hematopoietic Cell Transplanta-
tion). Ideally an examination of every sample before transplantation
to determine whether the cord blood cells contain abnormal clones
is suggested. In addition, long-term surveillance of stem cell trans-
plant recipients and donors is also required (Fig. 56.69).
Genetic Testing for Hematopoietic Cell Transplantation
Fig. 56.68 DETECTION OF ENGRAFTMENT AND RESIDUAL
DISEASE WITH FISH IN SEX-MISMATCHED HEMATOPOIETIC Testing consists of either variable number of tandem repeats/short
CELL TRANSPLANTATION. Metaphase and nondividing cell (blue) after tandem repeats PCR for detection of engraftment in patients with sex-
DAPI counterstaining, hybridized with X (large red) and Y (large green) for matched hematopoietic cell transplantation (HCT) or single-nucleotide
detection of engraftment and with ABL (small red) and BCR (small green) for polymorphism PCR, which has a high sensitivity. In sex-mismatched
detection of residual chronic myelogenous leukemia (top panel). Left nucleus HCT, both interphase FISH with XY probes and real-time quantitative
(bottom panel) shows a donor male (XY) cell origin and lack of BCR-ABL PCR for analysis of SRY gene on the Y chromosome can be used for
detection of engraftment. Detection of both engraftment and minimal
fusion. In contrast, a host female (XX), BCR-ABL fusion (yellow)–positive residual disease is best accomplished by FISH, simultaneously using
cell is shown on the right. Combination of XY FISH probes with diagnostic probes for XY and the explicit probe for diagnostic genetic defect, such
genomic markers is a powerful and fast FISH method for simultaneous as XY and BCR-ABL1.
detection of chimerism and minimal residual disease.