Page 1942 - Williams Hematology ( PDFDrive )
P. 1942
1916 Part XII: Hemostasis and Thrombosis Chapter 113: Molecular Biology and Biochemistry of the Coagulation Factors 1917
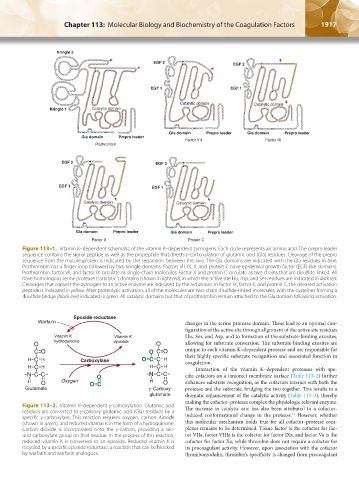
Kringle 2
EGF 2 EGF 2
EGF 1 EGF 1
Catalytic domain Catalytic domain
Kringle 1 Catalytic domain
Gla domain Prepro leader Gla domain Prepro leader
Gla domain Prepro leader
Factor VII Factor IX
Prothrombin
EGF 2 EGF 2
EGF 1 EGF 1
Catalytic domain Catalytic domain
Gla domain Prepro leader Gla domain Prepro leader
Factor X Protein C
Figure 113–1. Vitamin K–dependent schematic of the vitamin K–dependent zymogens. Each circle represents an amino acid. The prepro leader
sequence contains the signal peptide as well as the propeptide that directs γ-carboxylation of glutamic acid (Gla) residues. Cleavage of the prepro
sequence from the mature protein is indicated by the separation between the two. The Gla domains are indicated with the Gla residues in blue.
Prothrombin has a finger loop followed by two kringle domains. Factors VII, IX, X, and protein C have epidermal growth factor (EGF)-like domains.
Prothrombin, factor VII, and factor IX circulate as single-chain molecules. Factor X and protein C circulate as two chains that are disulfide linked. All
have homologous serine protease (“catalytic”) domains (shown in light red), in which the active site His, Asp, and Ser residues are indicated in dark red.
Cleavages that convert the zymogen to an active enzyme are indicated by the red arrows. In factor IX, factor X, and protein C, the released activation
peptide is indicated in yellow. After proteolytic activation, all of the molecules are two-chain disulfide-linked molecules, with the cysteines forming a
disulfide bridge (black line) indicated in green. All catalytic domains but that of prothrombin remain attached to the Gla domain following activation.
Epoxide reductase
Warfarin changes in the serine protease domain. These lead to an optimal con-
figuration of the active site through alignment of the active site residues
Vitamin K Vitamin K His, Ser, and Asp, and to formation of the substrate-binding exosites,
O hydroquinone epoxide O allowing for substrate conversion. The substrate-binding exosites are
C O O C O unique to each vitamin K–dependent protease and are responsible for
H C H Carboxylase OC C H their highly specific substrate recognition and associated function in
coagulation.
H C H H C H Interaction of the vitamin K–dependent proteases with spe-
N C C O N C C cific cofactors on a (anionic) membrane surface (Table 113–2) further
H O Oxygen OC H O enhances substrate recognition, as the cofactors interact with both the
γ
Glutamate -Carboxy- protease and the substrate, bridging the two together. This results in a
glutamate dramatic enhancement of the catalytic activity (Table 113–3), thereby
making the cofactor–protease complex the physiologic relevant enzyme.
Figure 113–2. Vitamin K–dependent γ-carboxylation. Glutamic acid The increase in catalytic rate has also been attributed to a cofactor-
residues are converted to γ-carboxy glutamic acid (Gla) residues by a induced conformational change in the protease. However, whether
11
specific γ-carboxylase. This reaction requires oxygen, carbon dioxide
(shown in green), and reduced vitamin K in the form of a hydroquinone. this molecular mechanism holds true for all cofactor–protease com-
Carbon dioxide is incorporated onto the γ-carbon, providing a sec- plexes remains to be determined. Tissue factor is the cofactor for fac-
ond carboxylate group on that residue. In the process of this reaction, tor VIIa, factor VIIIa is the cofactor for factor IXa, and factor Va is the
reduced vitamin K is converted to an epoxide. Reduced vitamin K is cofactor for factor Xa, while thrombin does not require a cofactor for
recycled by a specific epoxide reductase, a reaction that can be blocked its procoagulant activity. However, upon association with the cofactor
by warfarin and warfarin analogues. thrombomodulin, thrombin’s specificity is changed from procoagulant
Kaushansky_chapter 113_p1915-1948.indd 1917 9/21/15 2:39 PM