Page 1946 - Williams Hematology ( PDFDrive )
P. 1946
1920 Part XII: Hemostasis and Thrombosis Chapter 113: Molecular Biology and Biochemistry of the Coagulation Factors 1921
1 2 34 5 6 7 8 at this position dramatically reduces factor IX activity resulting in
Gene 13 kb hemophilia B. 46,47 An O-linked fucose (Ser61) and glucose (Ser63) are
found in the EGF 1 domain, in addition to several O-linked glycans in
the activation peptide (Thr159, Thr169, Thr172, and Thr179). Further
modification of the activation peptide includes N-linked glycosylation
mRNA 2.7 kb of Asn residues 157 and 167, which modulates the circulating levels of
factor IX. 48–50
Factor IX binds with high affinity to the extracellular matrix com-
ponent collagen IV via residue Lys5 in the Gla domain. 51,52 Although
factor IX variants incapable of collagen IV binding exhibit a greater
Exon 1 2 34 5 6 7 8
Protein Pro GLA EGF 1EGF 2 CR Catalytic domain recovery, collagen IV association generates an extravascular reservoir
of factor IX that enables prolonged action of factor IX at a hemostatic
Figure 113–7. Relationship of gene structure to protein structure in relevant region.
factor VII. The exons, introns, mRNA, and protein structure are as indi-
cated. The mRNA is 2.7 kb with a small 5′ untranslated region and a Factor IX Activation and Factor IXa Activity
relatively large 3′ untranslated region (light blue). In the protein, Pro indi- Limited proteolysis of factor IX at both Arg145 and Arg180 by either
cates the prepro leader sequence, GLA indicates the γ-carboxy glutamic the tissue factor–factor VIIa complex or factor XIa results in the release
acid (Gla) domain, and epidermal growth factor (EGF)-1 and -2, as well
as the serine protease (catalytic) domain, are indicated. CR indicates the of the activation peptide and generation of factor IXa (see Fig. 113–1).
connecting region that comprises the site of proteolytic activation. Cleavage at Arg180 generates factor IXaα, which displays catalytic activ-
ity toward synthetic substrates only, whereas fully active factor IXaβ is
formed following cleavage at Arg145. 53,54
although the reported prevalences vary between countries. Homozy- Factor IXa is a two-chain serine protease (Mr ≈45,000) that is com-
gotes and compound heterozygotes develop a hemorrhagic diathesis posed of a light chain of 145 residues (Mr ≈17,000) and the catalytic
that may vary from mild to severe. heavy chain of 235 residues (Mr ≈28,000), which are covalently linked
The human gene mutation database (http://www.hgmd.org) lists via a disulfide bond.
258 mutations in the factor VII gene. The majority of these are missense Factor IXa has a low catalytic efficiency as a result of impaired
mutations, but splicing and regulatory mutations also occur. Small access of substrates to the active site that results from steric and elec-
55
deletions account for almost 10 percent of the documented mutations. trostatic repulsion. Reversible interaction with the cofactor VIIIa on
Other gross gene abnormalities appear to be uncommon. anionic membranes and subsequent factor X binding leads to rear-
The factor VII gene harbors many common polymorphisms of rangement of the regions surrounding the active site and proteolytic
which three are notable: Arg353Gln in the catalytic domain, a 10-bp factor X activation.
insertion in the promotor region, and a variable number of 37 bp repeats The primary plasma inhibitor of factor IXa is the serpin anti-
in intron 7. The minor alleles of these polymorphisms are associated thrombin, and this inhibition is enhanced by heparin (see Table 113–4),
43
with decreased levels of factor VII and explain up to 30 percent of the which induces a conformational change in antithrombin that is required
variation in activated factor VII levels. Furthermore, the minor alleles for simultaneous active site and exosite interactions with factor IXa. 56
have been claimed to lower the risk of myocardial infarction. However,
this finding has not led to routine genotyping in the management of this Gene Structure and Variations
disorder. The relationship between factor VII levels, factor VII polymor- The gene encoding factor IX (F9) is located on chromosome Xq27.1 and
57
phisms, and venous thrombosis has not been established with certainty. covers nearly 25 kb. It is divided into eight exons from which a mature
mRNA molecule is transcribed with an ultimate length of 2802 bases
(Fig. 113–8).
FACTOR IX
Factor IX was originally reported in 1952 as “Christmas factor,” named
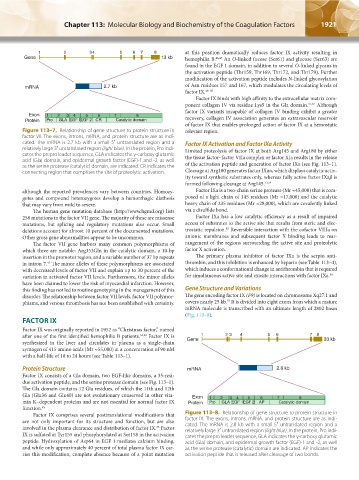
after one of the first identified hemophilia B patients. 34,44 Factor IX is 1 2 3 4 5 6 7 8
synthesized in the liver and circulates in plasma as a single-chain Gene 33 kb
zymogen of 415 amino acids (Mr ≈55,000) at a concentration of 90 nM
with a half-life of 18 to 24 hours (see Table 113–1).
Protein Structure mRNA 2.8 kb
Factor IX consists of a Gla domain, two EGF-like domains, a 35-resi-
due activation peptide, and the serine protease domain (see Fig. 113–1).
The Gla domain contains 12 Gla residues, of which the 11th and 12th
Gla (Glu36 and Glu40) are not evolutionary conserved in other vita- Exon
3
8
4
6
5
min K–dependent proteins and are not essential for normal factor IX Protein 1 Pro 2 GLA EGF 1EGF 2 AP 7 Catalytic domain
function. 45
Factor IX comprises several posttranslational modifications that Figure 113–8. Relationship of gene structure to protein structure in
are not only important for its structure and function, but are also factor IX. The exons, introns, mRNA, and protein structure are as indi-
involved in the plasma clearance and distribution of factor IX. Factor cated. The mRNA is 2.8 kb with a small 5′ untranslated region and a
35
relatively large 3′ untranslated region (light blue). In the protein, Pro indi-
IX is sulfated at Tyr155 and phosphorylated at Ser158 in the activation cates the prepro leader sequence, GLA indicates the γ-carboxy glutamic
peptide. Hydroxylation of Asp64 in EGF 1 mediates calcium binding, acid (Gla) domain, and epidermal growth factor (EGF)-1 and -2, as well
and while only approximately 40 percent of total plasma factor IX car- as the serine protease (catalytic) domain are indicated. AP indicates the
ries this modification, complete absence because of a point mutation activation peptide that is released after cleavage of two bonds.
Kaushansky_chapter 113_p1915-1948.indd 1921 9/21/15 2:39 PM