Page 1943 - Williams Hematology ( PDFDrive )
P. 1943
1918 Part XII: Hemostasis and Thrombosis Chapter 113: Molecular Biology and Biochemistry of the Coagulation Factors 1919
TABLE 113–3. Cofactor Enhancement of Serine Protease
Activity
Cofactor-Protease * Fold Increase †
7 Ca ++
TM-Thrombin 11,000
TF-VIIa 31,000
VIIIa-IXa 9,000,000
Va-Xa 390,000
TF, tissue factor; TM, thrombomodulin.
GLA residue * Macromolecular enzyme complexes assembled in the presence of
Hydrophobic residue anionic phospholipids and calcium.
† Relative rates of enzymatic activity represent fold increase of the
Lipid membrane
reaction rate (k /Km) observed for the cofactor-protease complex
cat
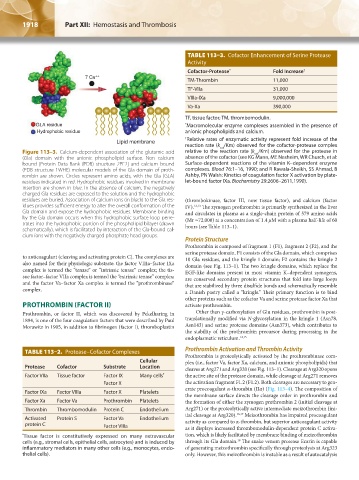
Figure 113–3. Calcium-dependent association of the glutamic acid relative to the reaction rate (k /Km) observed for the protease in
cat
(Gla) domain with the anionic phospholipid surface. Non–calcium absence of the cofactor (see KG Mann, ME Nesheim, WR Church, et al:
bound [Protein Data Bank (PDB) structure 2PF2] and calcium bound Surface-dependent reactions of the vitamin K–dependent enzyme
(PDB structure 1WHE) molecular models of the Gla domain of proth- complexes. Blood 76:1–16, 1990; and R Rawala-Sheikh, SS Ahmad, B
rombin are shown. Circles represent amino acids, with the Gla (GLA) Ashby, PN Walsh: Kinetics of coagulation factor X activation by plate-
residues indicated in red. Hydrophobic residues involved in membrane let-bound factor IXa. Biochemistry 29:2606–2611,1990).
insertion are shown in blue. In the absence of calcium, the negatively
charged Gla residues are exposed to the solution and the hydrophobic
residues are buried. Association of calcium ions (in black) to the Gla res- (thrombokinase, factor III, now tissue factor), and calcium (factor
idues provides sufficient energy to alter the overall conformation of the IV). 12,13 The zymogen prothrombin is primarily synthesized in the liver
Gla domain and expose the hydrophobic residues. Membrane binding and circulates in plasma as a single-chain protein of 579 amino acids
by the Gla domain occurs when this hydrophobic surface loop pene- (Mr ≈72,000) at a concentration of 1.4 μM with a plasma half-life of 60
trates into the hydrophobic portion of the phospholipid bilayer (drawn hours (see Table 113–1).
schematically), which is facilitated by interaction of the Gla-bound cal-
cium ions with the negatively charged phosphate head groups.
Protein Structure
Prothrombin is composed of fragment 1 (F1), fragment 2 (F2), and the
serine protease domain. F1 consists of the Gla domain, which comprises
to anticoagulant (cleaving and activating protein C). The complexes are 10 Gla residues, and the kringle 1 domain; F2 contains the kringle 2
also named for their physiologic substrate: the factor VIIIa–factor IXa domain (see Fig. 113–1). The two kringle domains, which replace the
complex is termed the “tenase” or “intrinsic tenase” complex; the tis- EGF-like domains present in most vitamin K–dependent zymogens,
sue factor–factor VIIa complex is termed the “extrinsic tenase” complex; are conserved secondary protein structures that fold into large loops
and the factor Va–factor Xa complex is termed the “prothrombinase” that are stabilized by three disulfide bonds and schematically resemble
complex.
a Danish pastry called a “kringle.” Their primary function is to bind
other proteins such as the cofactor Va and serine protease factor Xa that
PROTHROMBIN (FACTOR II) activate prothrombin.
Prothrombin, or factor II, which was discovered by Pekelharing in Other than γ-carboxylation of Glu residues, prothrombin is post-
1894, is one of the four coagulation factors that were described by Paul translationally modified via N-glycosylation in the kringle 1 (Asn78,
Morawitz in 1905, in addition to fibrinogen (factor I), thromboplastin Asn143) and serine protease domains (Asn373), which contributes to
the stability of the prothrombin precursor during processing in the
endoplasmatic reticulum. 14,15
TABLE 113–2. Protease–Cofactor Complexes Prothrombin Activation and Thrombin Activity
Prothrombin is proteolytically activated by the prothrombinase com-
Cellular plex (i.e., factor Va, factor Xa, calcium, and anionic phospholipids) that
Protease Cofactor Substrate Location cleaves at Arg271 and Arg320 (see Fig. 113–1). Cleavage at Arg320 opens
Factor VIIa Tissue factor Factor IX Many cells * the active site of the protease domain, while cleavage at Arg271 removes
Factor X the activation fragment F1.2 (F1.2). Both cleavages are necessary to gen-
erate procoagulant α-thrombin (IIα) (Fig. 113–4). The composition of
Factor IXa Factor VIIIa Factor X Platelets
the membrane surface directs the cleavage order in prothrombin and
Factor Xa Factor Va Prothrombin Platelets the formation of either the zymogen prethrombin 2 (initial cleavage at
Thrombin Thrombomodulin Protein C Endothelium Arg271) or the proteolytically active intermediate meizothrombin (ini-
tial cleavage at Arg320). 16,17 Meizothrombin has impaired procoagulant
Activated Protein S Factor Va Endothelium
protein C Factor VIIIa activity as compared to α-thrombin, but superior anticoagulant activity
as it displays increased thrombomodulin-dependent protein C activa-
* Tissue factor is constitutively expressed on many extravascular tion, which is likely facilitated by membrane binding of meizothrombin
18
cells (e.g., stromal cells, epithelial cells, astrocytes) and is induced by through its Gla domain. The snake venom protease Ecarin is capable
inflammatory mediators in many other cells (e.g., monocytes, endo- of generating meizothrombin specifically through proteolysis at Arg323
thelial cells). only. However, this meizothrombin is instable as a result of autocatalysis
Kaushansky_chapter 113_p1915-1948.indd 1918 9/21/15 2:39 PM