Page 265 - Williams Hematology ( PDFDrive )
P. 265
240 Part IV: Molecular and Cellular Hematology Chapter 16: Cell-Cycle Regulation and Hematologic Disorders 241
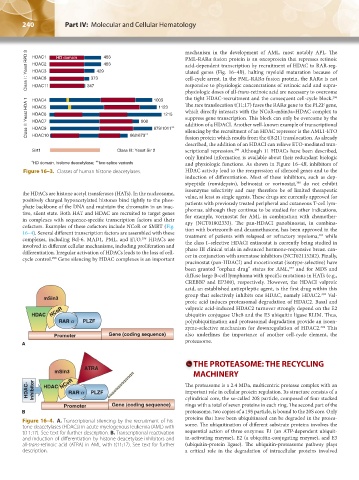
Class I : Yeast RPD 3 HDAC1 HD domain 373 429 483 mechanism in the development of AML, most notably APL. The
PML-RARα fusion protein is an oncoprotein that represses retinoic
483
HDAC2
acid-dependent transcription by recruitment of HDAC to RAR-reg-
HDAC3
ulated genes (Fig. 16–4B), halting myeloid maturation because of
cell-cycle arrest. In the PML-RARα fusion protein, the RARα is not
HDAC8
responsive to physiologic concentrations of retinoic acid and supra-
347
HDAC11
physiologic doses of all-trans-retinoic acid are necessary to overcome
the tight HDAC-recruitment and the consequent cell-cycle block.
296
HDAC4
Class II: Yeast HDA 1 HDAC5 808 879/1011 ∗∗ which directly interacts with the NCoR–mSin3a–HDAC complex to
1003
The rare translocation t(11;17) fuses the RARα gene to the PLZF gene,
1123
1215
HDAC6
suppress gene transcription. This block can only be overcome by the
HDAC7
addition of a HDACI. Another well-known example of transcriptional
HDAC9
silencing by the recruitment of an HDAC repressor is the AML1-ETO
∗∗
HDAC10
662/673
fusion protein which results from the t(8;21) translocation. As already
described, the addition of an HDACI can relieve ETO-mediated tran-
299
Sirt1 Class III: Yeast Sir 2 scriptional repression. Although 11 HDACs have been described,
only limited information is available about their redundant biologic
∗ HD domain, histone deacetylase; two-splice variants and physiologic functions. As shown in Figure 16–4B, inhibitors of
∗∗
Figure 16–3. Classes of human histone deacetylases. HDAC activity lead to the reexpression of silenced genes and to the
induction of differentiation. Most of these inhibitors, such as dep-
300
sipeptide (romidepsin), belinostat or vorinostat, do not exhibit
isoenzyme selectivity and may therefore be of limited therapeutic
the HDACs are histone acetyl transferases (HATs). In the nucleosome,
positively charged hypoacetylated histones bind tightly to the phos- value, at least as single agents. These drugs are currently approved for
phate backbone of the DNA and maintain the chromatin in an inac- patients with previously treated peripheral and cutaneous T-cell lym-
tive, silent state. Both HAT and HDAC are recruited to target genes phomas, although they continue to be studied for other indications,
in complexes with sequence-specific transcription factors and their for example, vorinostat for AML in combination with chemother-
cofactors. Examples of these cofactors include NCoR or SMRT (Fig. apy (NCT01802333). The pan-HDACI panobinostat, in combina-
16–4). Several different transcription factors are assembled with these tion with bortezomib and dexamethasone, has been approved in the
301
complexes, including Bcl-6, MAD1, PML, and ETO. HDACs are treatment of patients with relapsed or refractory myeloma, while
296
involved in different cellular mechanisms, including proliferation and the class I–selective HDACI entinostat is currently being studied in
differentiation. Irregular activation of HDACs leads to the loss of cell- phase III clinical trials in advanced hormone-responsive breast can-
cycle control. Gene silencing by HDAC complexes is an important cer in conjunction with aromatase inhibitors (NCT02115282). Finally,
298
pracinostat (pan-HDACI) and mocetinostat (isotype-selective) have
147
been granted “orphan drug” status for AML, and for MDS and
diffuse large B-cell lymphoma with specific mutations in HATs (e.g.,
CREBBP and EP300), respectively. However, the HDACI valproic
acid, an established antiepileptic agent, is the first drug within this
group that selectively inhibits one HDAC, namely HDAC2. Val-
302
proic acid induces proteasomal degradation of HDAC2. Basal and
valproic acid-induced HDAC2 turnover strongly depend on the E2
ubiquitin conjugase Ubc8 and the E3 ubiquitin ligase RLIM. Thus,
polyubiquitination and proteasomal degradation provide an isoen-
302
zyme-selective mechanism for downregulation of HDAC2. This
also underlines the importance of another cell-cycle element, the
proteasome.
A
THE PROTEASOME: THE RECYCLING
MACHINERY
The proteasome is a 2.4 MDa, multicentric protease complex with an
important role in cellular protein regulation. Its structure consists of a
cylindrical core, the so-called 20S particle, composed of four stacked
rings with a total of seven proteins in each ring. The second part of the
B proteasome, two copies of a 19S particle, is bound to the 20S core. Only
proteins that have been ubiquitinated can be degraded in the protea-
Figure 16–4. A. Transcriptional silencing by the recruitment of his-
tone deacetylases (HDACs) in acute myelogenous leukemia (AML) with some. The ubiquitination of different substrate proteins involves the
t(11;17). See text for further description. B. Transcriptional reactivation sequential action of three enzymes: E1 (an ATP-dependent ubiquit-
and induction of differentiation by histone deacetylase inhibitors and in-activating enzyme), E2 (a ubiquitin-conjugating enzyme), and E3
all-trans-retinoic acid (ATRA) in AML with t(11;17). See text for further (ubiquitin-protein ligase). The ubiquitin-proteasome pathway plays
description. a critical role in the degradation of intracellular proteins involved
Kaushansky_chapter 16_p0213-0246.indd 240 9/18/15 11:58 PM