Page 78 - Clinical Immunology_ Principles and Practice ( PDFDrive )
P. 78
CHaPter 4 Antigen Receptor Genes, Gene Products, and Coreceptors 63
H
H
H
Ig H chain locus V (39) D (27) J (6) C (9)
H
Chr. 14q32.2
V3-74 V6-1 D1-1 D7-27 CD252 123456 Cµ Cδ Cγ3 Cγ1 Cα1 Cγ2 Cγ4 Cε Cα2
5’ S S S S S S S S 3’
Ig κ chain locus V (40) J (5) C (1)
κ
κ
κ
Chr. 2p 1.2
3D-7 4.1 12345 Cκ κde
5’ 3’
iEκ 3’E κ
Ig λ chain locus V (40) J - C cluster (4)
λ
λ λ
Chr. 22q 11.2
4.69 3.1 Jλ1 Cλ1 Jλ2 Cλ2 Jλ3 Cλ3 Jλ7 Cλ7
5’ 3’
3’E κ
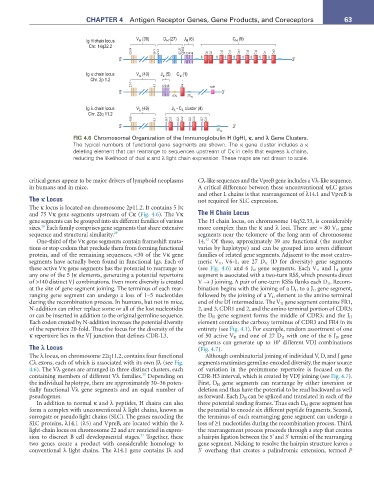
FIG 4.6 Chromosomal Organization of the Immunoglobulin H (IgH), κ, and λ Gene Clusters.
The typical numbers of functional gene segments are shown. The κ gene cluster includes a κ
deleting element that can rearrange to sequences upstream of Cκ in cells that express λ chains,
reducing the likelihood of dual κ and λ light chain expression. These maps are not drawn to scale.
critical genes appear to be major drivers of lymphoid neoplasms Cλ-like sequences and the VpreB gene includes a Vλ-like sequence.
in humans and in mice. A critical difference between these unconventional ψLC genes
and other L chains is that rearrangement of λ14.1 and VpreB is
The κ Locus not required for SLC expression.
The κ locus is located on chromosome 2p11.2. It contains 5 Jκ
and 75 Vκ gene segments upstream of Cκ (Fig. 4.6). The Vκ The H Chain Locus
gene segments can be grouped into six different families of various The H chain locus, on chromosome 14q32.33, is considerably
28
sizes. Each family comprises gene segments that share extensive more complex than the κ and λ loci. There are ≈ 80 V H gene
sequence and structural similarity. 29 segments near the telomere of the long arm of chromosome
32
One-third of the Vκ gene segments contain frameshift muta- 14. Of these, approximately 39 are functional (the number
tions or stop codons that preclude them from forming functional varies by haplotype) and can be grouped into seven different
protein, and of the remaining sequences, <30 of the Vκ gene families of related gene segments. Adjacent to the most centro-
segments have actually been found in functional Igs. Each of meric V H , V6-1, are 27 D H (D for diversity) gene segments
these active Vκ gene segments has the potential to rearrange to (see Fig. 4.6) and 6 J H gene segments. Each V H and J H gene
any one of the 5 Jκ elements, generating a potential repertoire segment is associated with a two-turn RSS, which prevents direct
of >140 distinct VJ combinations. Even more diversity is created V → J joining. A pair of one-turn RSSs flanks each D H . Recom-
at the site of gene segment joining. The terminus of each rear- bination begins with the joining of a D H to a J H gene segment,
ranging gene segment can undergo a loss of 1–5 nucleotides followed by the joining of a V H element to the amino terminal
during the recombination process. In humans, but not in mice, end of the DJ intermediate. The V H gene segment contains FR1,
N-addition can either replace some or all of the lost nucleotides 2, and 3, CDR1 and 2, and the amino terminal portion of CDR3;
or can be inserted in addition to the original germline sequence. the D H gene segment forms the middle of CDR3; and the J H
Each codon created by N-addition increases the potential diversity element contains the carboxy terminus of CDR3 and FR4 in its
of the repertoire 20-fold. Thus the focus for the diversity of the entirety (see Fig. 4.1). For example, random assortment of one
κ repertoire lies in the VJ junction that defines CDR-L3. of 50 active V H and one of 27 D H with one of the 6 J H gene
4
segments can generate up to 10 different VDJ combinations
The λ Locus (Fig. 4.7).
The λ locus, on chromosome 22q11.2, contains four functional Although combinatorial joining of individual V, D, and J gene
Cλ exons, each of which is associated with its own Jλ (see Fig. segments maximizes germline-encoded diversity, the major source
4.6). The Vλ genes are arranged in three distinct clusters, each of variation in the preimmune repertoire is focused on the
30
containing members of different Vλ families. Depending on CDR-H3 interval, which is created by VDJ joining (see Fig. 4.7).
the individual haplotype, there are approximately 30–36 poten- First, D H gene segments can rearrange by either inversion or
tially functional Vλ gene segments and an equal number of deletion and thus have the potential to be read backward as well
pseudogenes. as forward. Each D H can be spliced and translated in each of the
In addition to normal κ and λ peptides, H chains can also three potential reading frames. Thus each D H gene segment has
form a complex with unconventional λ light chains, known as the potential to encode six different peptide fragments. Second,
surrogate or pseudo light chains (SLC). The genes encoding the the terminus of each rearranging gene segment can undergo a
SLC proteins, λ14.1 (λ5) and VpreB, are located within the λ loss of ≥1 nucleotides during the recombination process. Third,
light-chain locus on chromosome 22 and are restricted in expres- the rearrangement process proceeds through a step that creates
31
sion to discreet B cell developmental stages. Together, these a hairpin ligation between the 5’ and 3’ termini of the rearranging
two genes create a product with considerable homology to gene segment. Nicking to resolve the hairpin structure leaves a
conventional λ light chains. The λ14.1 gene contains Jλ and 3’ overhang that creates a palindromic extension, termed P