Page 679 - Clinical Hematology_ Theory _ Procedures ( PDFDrive )
P. 679
CHAPTER 31 ■ Molecular Diagnostic Techniques and Applications 663
1. emplate preparation
2. Sequencing and imaging
3. Data analysis
emplate Preparation. T is step consists o building a DNA
or complementary DNA (cDNA) library and ampli cation
o that library. A sequencing library is constructed by
ragmenting the DNA or cDNA specimen and attaching
adapter sequences, synthetic oligonucleotides o a known
sequence, to the ends o the DNA ragments. A constructed
library is clonally ampli ed in preparation or sequencing.
Ampli cation o single library ragment on microbeads is
unique to the One ouch system; bridge ampli cation is
used to orm template clusters on a f ow cell by the Illumina
FIGURE 31.11 FISH technique demonstrates trisomy 12 in a
case o CLL with prolymphocytoid trans ormation. (From Sun . system.
Flow Cytometry, Immunohistochemistry, and Molecular Genetics Sequencing and Imaging. For the next step, the two com-
or Hematologic Neoplasms, 2nd ed, Philadelphia, PA: Lippincott mercial systems rely on sequencing by synthesis. T e library
Williams & Wilkins, 2012.) ragments act as a template rom which a new DNA rag-
ment is synthesized. Sequencing occurs through a cycle o
washing and f ooding the ragments with the known nucle-
cally used assay is based on DNA sequencing. Commercial otides in a sequential order. As nucleotides incorporate
panels available through Ion orrent PGM One ouch into a growing DNA strand, they are digitally recorded as
system and Illumina are popular or design own panels based sequence. One system, PGM, does semiconductor sequenc-
on genes o interest. NCS permits analysis o all the exomes ing that relies on detection o pH changes induced by the
or even entire genes. T is has led to expansion o the number release o a hydrogen ion upon the incorporation o a nucle-
o genes that can be analyzed at one time and identi cation otide into a growing strand o DNA. T e Illumina MiSeq
o patients at risk. relies on detection o f uorescence generated by the incorpo-
Steps in NGS ration o f uorescently labeled nucleotides into the growing
Each NGS plat orm is unique in how sequencing is strand o DNA.
accomplished, but the generalized sequencing protocol or Data Analysis. T e nal step a er sequencing is complete
the two commercially available NGS plat orms (Ion orrent is that raw sequence data must undergo several analysis
PGM One ouch system and Illumina) includes steps. Preprocessing o data removes adapter sequences and
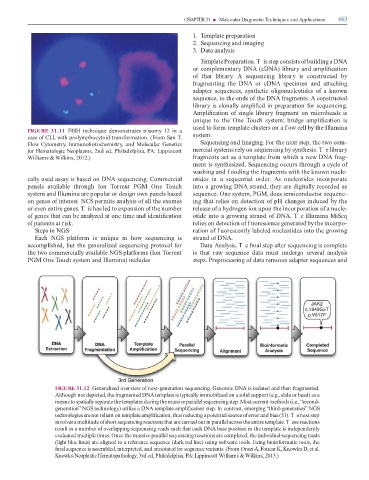
FIGURE 31.12 Generalized overview o next-generation sequencing. Genomic DNA is isolated and then ragmented.
Although not depicted, the ragmented DNA template is typically immobilized on a solid support (e.g., slide or bead) as a
means to spatially separate the templates during the massive parallel sequencing step. Most current methods (i.e., “second-
generation” NGS technology) utilize a DNA template ampli cation step. In contrast, emerging “third-generation” NGS
technologies are not reliant on template ampli cation, thus reducing a potential source o error and bias (51). T e next step
involves a multitude o short sequencing reactions that are carried out in parallel across the entire template. T ese reactions
result in a number o overlapping sequencing reads such that each DNA base position in the template is independently
evaluated multiple times. Once the massive parallel sequencing reactions are completed, the individual sequencing reads
(light blue lines) are aligned to a re erence sequence (dark red line) using so ware tools. Using bioin ormatic tools, the
nal sequence is assembled, interpreted, and annotated or sequence variants. (From Orazi A, Foucar K, Knowles D, et al.
Knowles Neoplastic Hematopathology, 3rd ed, Philadelphia, PA: Lippincott Williams & Wilkins, 2013.)