Page 678 - Clinical Hematology_ Theory _ Procedures ( PDFDrive )
P. 678
662 PART 8 ■ Fundamentals of Hematological Analysis
A
gene A (normal) CC TG AGG
BOX 31.4
Ms t II site
gene S (sickle) CC TG T GG
Strengths and Limitations of FISH
(no Ms t II site)
STRENGTHS OF FISH
B Restriction site ■ Is better than PCR or deletions and inversions
absent in sickle-cell β-globin ■ Is the next generation o cytogenetic techniques a er
β-globin gene
banding
Ms t II Ms t II Ms t II ■ Can be correlated with morphology
■ Can be used to assess DNA or mRNA
■ Can be per ormed on metaphase spreads but can also
gene A 1.1kb be done on para n block sections
gene S 1.3kb
LIMITATIONS OF FISH
■ Problems in tissue that has been decalci ed or not xed
C in ormalin
Southern blot ■ Requires a f uorescent microscope
of DNA cut with Probes must be speci cally designed
Ms t II and β (1.3kb) ■
S
hybridized ■ Can only be used to detect the presence or absence o
with β-globin β (1.1kb) previously identi ed chromosomal aberrations
A
probe ■ Not good as PCR or very small mutations
■ Labor intensive
■ issue that is xed cannot be cultured or metaphase
Sickle-cell control
Normal control spreads
Carrier
Modi ed rom Heriot K. Welcome to the beginning: molecular pathol-
Affected individual ogy or the community hospital pathologist and medical technologist,
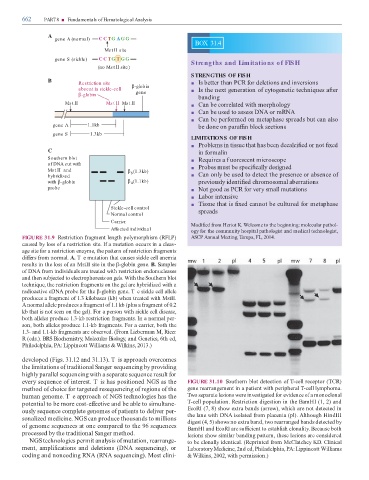
FIGURE 31.9 Restriction ragment length polymorphism (RFLP) ASCP Annual Meeting, ampa, FL, 2014.
caused by loss o a restriction site. I a mutation occurs in a cleav-
age site or a restriction enzyme, the pattern o restriction ragments
di ers rom normal. A. T e mutation that causes sickle cell anemia
results in the loss o an MstII site in the β-globin gene. B. Samples
o DNA rom individuals are treated with restriction endonucleases
and then subjected to electrophoresis on gels. With the Southern blot
technique, the restriction ragments on the gel are hybridized with a
radioactive cDNA probe or the β-globin gene. T e sickle cell allele
produces a ragment o 1.3 kilobases (kb) when treated with MstII.
A normal allele produces a ragment o 1.1 kb (plus a ragment o 0.2
kb that is not seen on the gel). For a person with sickle cell disease,
both alleles produce 1.3-kb restriction ragments. In a normal per-
son, both alleles produce 1.1-kb ragments. For a carrier, both the
1.3- and 1.1-kb ragments are observed. (From Lieberman M, Ricer
R (eds.). BRS Biochemistry, Molecular Biology, and Genetics, 6th ed,
Philadelphia, PA: Lippincott Williams & Wilkins, 2013.)
developed (Figs. 31.12 and 31.13). T is approach overcomes
the limitations o traditional Sanger sequencing by providing
highly parallel sequencing with a separate sequence result or
every sequence o interest. T is has positioned NGS as the FIGURE 31.10 Southern blot detection o -cell receptor ( CR)
method o choice or targeted resequencing o regions o the gene rearrangement in a patient with peripheral -cell lymphoma.
human genome. T e approach o NGS technologies has the wo separate lesions were investigated or evidence o a monoclonal
potential to be more cost-e ective and be able to simultane- -cell population. Restriction digestion in the BamHI (1, 2) and
ously sequence complete genomes o patients to deliver per- EcoRI (7, 8) show extra bands (arrow), which are not detected in
sonalized medicine. NGS can produce thousands to millions the lane with DNA isolated rom placenta (pl). Although HindIII
digest (4, 5) shows no extra band, two rearranged bands detected by
o genome sequences at one compared to the 96 sequences BamHI and EcoRI are su cient to establish clonality. Because both
processed by the traditional Sanger method. lesions show similar banding pattern, these lesions are considered
NGS technologies permit analysis o mutation, rearrange- to be clonally identical. (Reprinted rom McClatchey KD. Clinical
ment, ampli cations and deletions (DNA sequencing), or Laboratory Medicine, 2nd ed, Philadelphia, PA: Lippincott Williams
coding and noncoding RNA (RNA sequencing). Most clini- & Wilkins, 2002, with permission.)