Page 903 - Hematology_ Basic Principles and Practice ( PDFDrive )
P. 903
786 Part VII Hematologic Malignancies
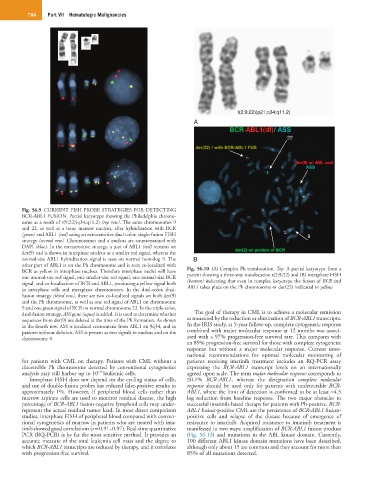
t(2;9;22)(p21;q34;q11.2)
A
22
Fig. 56.9 CURRENT FISH PROBE STRATEGIES FOR DETECTING
BCR-ABL1 FUSION. Partial karyotype showing the Philadelphia chromo-
some as a result of t(9;22)(q34;q11.2) (top row). The same chromosomes 9
and 22, as well as a bone marrow nucleus, after hybridization with BCR
(green) and ABL1 (red) using an extrasensitive dual-color, single-fusion FISH 9
strategy (second row). Chromosomes and a nucleus are counterstained with
DAPI (blue). In the extrasensitive strategy, a part of ABL1 (red) remains on
der(9) and is shown in interphase nucleus as a smaller red signal, whereas the
normal-size ABL1 hybridization signal is seen on normal homolog 9. The B
other part of ABL1 is on the Ph chromosome and is seen co-localized with
BCR as yellow in interphase nucleus. Therefore interphase nuclei will have Fig. 56.10 (A) Complex Ph translocation. Top: A partial karyotype from a
one normal-size red signal, one smaller-size red signal, one normal-size BCR patient showing a three-way translocation t(2;9;22) and (B) metaphase FISH
signal, and co-localization of BCR and ABL1, producing a yellow signal both (bottom) indicating that even in complex karyotype the fusion of BCR and
in interphase cells and metaphase chromosomes. In the dual-color, dual- ABL1 takes place on the Ph chromosome or der(22) indicated in yellow.
fusion strategy (third row), there are two co-localized signals on both der(9)
and the Ph chromosome, as well as one red signal of ABL1 on chromosome
9 and one green signal of BCR on normal chromosome 22. In the triple-color,
dual-fusion strategy, ASS gene (aqua) is added. It is used to determine whether The goal of therapy in CML is to achieve a molecular remission
sequences from der(9) are deleted at the time of the Ph formation. As shown as measured by the reduction or elimination of BCR-ABL1 transcripts.
in the fourth row, ASS is localized centromeric from ABL1 on 9q34, and in In the IRIS study, at 5-year follow-up, complete cytogenetic response
patients without deletion, ASS is present as two signals in nucleus and on the combined with major molecular response at 12 months was associ-
chromosome 9. ated with a 97% progression-free survival rate. This compares with
an 89% progression-free survival for those with complete cytogenetic
response but without a major molecular response. Current inter-
national recommendations for optimal molecular monitoring of
for patients with CML on therapy. Patients with CML without a patients receiving imatinib treatment includes an RQ-PCR assay
discernible Ph chromosome detected by conventional cytogenetics expressing the BCR-ABL1 transcript levels on an internationally
10
analysis may still harbor up to 10 leukemic cells. agreed upon scale. The term major molecular response corresponds to
Interphase FISH does not depend on the cycling status of cells, ≤0.1% BCR-ABL1, whereas the designation complete molecular
and use of double-fusion probes has reduced false-positive results to response should be used only for patients with undetectable BCR-
approximately 1%. However, if peripheral blood cells rather than ABL1, where the limit of detection is confirmed to be at least ≈4.5
marrow aspirate cells are used to monitor residual disease, the high log reduction from baseline response. The two major obstacles to
percentage of BCR-ABL1 fusion-negative lymphoid cells may under- successful imatinib-based therapy for patients with Ph-positive, BCR-
represent the actual residual tumor load. In most direct comparison ABL1 fusion-positive CML are the persistence of BCR-ABL1 fusion-
studies, interphase FISH of peripheral blood compared with conven- positive cells and relapse of the disease because of emergence of
tional cytogenetics of marrow in patients who are treated with ima- resistance to imatinib. Acquired resistance to imatinib treatment is
tinib showed good correlations (r = 0.91–0.97). Real-time quantitative manifested in two ways: amplification of BCR-ABL1 fusion product
PCR (RQ-PCR) is by far the most sensitive method. It provides an (Fig. 56.13) and mutations in the ABL kinase domain. Currently,
accurate measure of the total leukemia cell mass and the degree to 100 different ABL1 kinase domain mutations have been described,
which BCR-ABL1 transcripts are reduced by therapy, and it correlates although only about 15 are common and they account for more than
with progression-free survival. 85% of all mutations detected.