Page 907 - Hematology_ Basic Principles and Practice ( PDFDrive )
P. 907
790 Part VII Hematologic Malignancies
1 2 3 4 5
8 8 der(8) 21 der(21)
6 7 8 9 10 11 12
9 der(9) 22 Ph
13 14 15 16 17 18
15 15 15
19 20 21 22 X Y
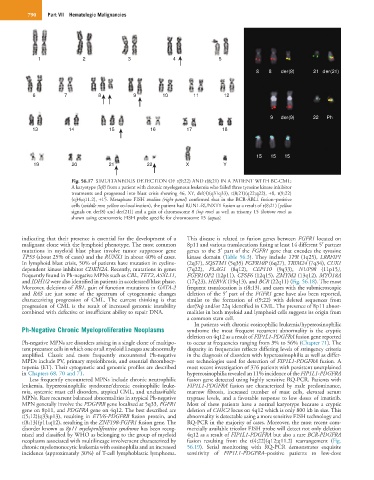
Fig. 56.17 SIMULTANEOUS DETECTION OF t(9;22) AND t(8;21) IN A PATIENT WITH BC-CML:
A karyotype (left) from a patient with chronic myelogenous leukemia who failed three tyrosine kinase inhibitor
treatments and progressed into blast crisis showing 46, XY, del(4)(q31q33), t(8;21)(q22;q22), +8, t(9;22)
(q34;q11.2), +15. Metaphase FISH studies (right panel) confirmed that in the BCR-ABL1 fusion–positive
cells (middle row, yellow co-localization), the patient had RUN1-RUNXT1 fusion as a result of t(8;21) [yellow
signals on der(8) and der(21)] and a gain of chromosome 8 (top row) as well as trisomy 15 (bottom row) as
shown using centromeric FISH probe specific for chromosome 15 (aqua).
indicating that their presence is essential for the development of a This disease is related to fusion genes between FGFR1 located on
malignant clone with the lymphoid phenotype. The most common 8p11 and various translocations fusing at least 14 different 5′ partner
mutations in myeloid blast phase involve tumor suppressor gene genes to the 3′ part of the FGFR1 gene that encodes the tyrosine
TP53 (about 25% of cases) and the RUNX1 in about 40% of cases. kinase domain (Table 56.3). They include TPR (1q25), LRRFIP1
In lymphoid blast crisis, 50% of patients have mutation in cycline- (2q37), SQSTM1 (5q35) FGFR10P (6q27), TRIM24 (7q34), CUX1
dependent kinase inhibitor CDKN2A. Recently, mutations in genes (7q22), PLAG1 (8q12), CEP110 (9q33), NUP98 (11p15),
frequently found in Ph-negative MPNs such as CBL, TET2, ASXL11, FGFR1OP2 (12p11), CPSF6 (12q15), ZMYM2 (13q12), MYO18A
and IDH1/2 were also identified in patients in accelerated/blast phase. (17q23), HERVK (19q13), and BCR (22q11) (Fig. 56.18). The most
Moreover, deletions of RB1, gain of function mutations in GATA-2 frequent translocation is t(8;13), and cases with the submicroscopic
and RAS are just some of the spectrum of cytogenomic changes deletion of the 5′ part of the FGFR1 gene have also been reported,
characterizing progression of CML. The current thinking is that similar to the formation of t(9;22) with deleted sequences from
progression of CML is the result of increased genomic instability der(9q) and/or 22q identified in CML. The presence of 8p11 abnor-
combined with defective or insufficient ability to repair DNA. malities in both myeloid and lymphoid cells suggests its origin from
a common stem cell.
In patients with chronic eosinophilic leukemia/hypereosinophilic
Ph-Negative Chronic Myeloproliferative Neoplasms syndrome the most frequent recurrent abnormality is the cryptic
deletion on 4q12 as a result of FIP1L1-PDGFRA fusion gene reported
Ph-negative MPNs are disorders arising in a single clone of multipo- to occur at frequencies ranging from 3% to 56% (Chapter 71). The
tent precursor cells in which one or all myeloid lineages are abnormally disparity in frequencies reflects differing levels of stringency criteria
amplified. Classic and more frequently encountered Ph-negative in the diagnosis of disorders with hypereosinophilia as well as differ-
MPDs include PV, primary myelofibrosis, and essential thrombocy- ent technologies used for detection of FIP1L1-PDGFRA fusion. A
topenia (ET). Their cytogenetic and genomic profiles are described most recent investigation of 376 patients with persistent unexplained
in Chapters 68, 70 and 71. hypereosinophilia revealed an 11% incidence of the FIP1L1-PDGFRA
Less frequently encountered MPNs include chronic neutrophilic fusion gene detected using highly sensitive RQ-PCR. Patients with
leukemia, hypereosinophilic syndrome/chronic eosinophilic leuke- FIP1L1-PDGFRA fusion are characterized by male predominance,
mia, systemic mast cell disorders, atypical CML, and unclassifiable marrow fibrosis, increased number of mast cells, elevated serum
MPNs. Rare recurrent balanced abnormalities in atypical Ph-negative tryptase levels, and a favorable response to low doses of imatinib.
MPN generally involve the PDGFRB gene localized at 5q33, FGFR1 Most of these patients have a normal karyotype because a cryptic
gene on 8p11, and PDGFRA gene on 4q12. The best described are deletion of CHIC2 locus on 4q12 which is only 800 kb in size. This
t(5;12)(q33;p13), resulting in ETV6-PDGFRB fusion protein, and abnormality is detectable using a more sensitive FISH technology and
t(8;13)(p11;q12), resulting in the ZNF198-FGFR1 fusion gene. The RQ-PCR in the majority of cases. Moreover, the most recent com-
disorder known as 8p11 myeloproliferative syndrome has been recog- mercially available tricolor FISH probe will detect not only deletion
nized and classified by WHO as belonging to the group of myeloid 4q12 as a result of FIP1L1-PDGFRA but also a rare BCR-PDGFRA
neoplasms associated with multilineage involvement characterized by fusion resulting from the t(4;22)(q12;q11.2) rearrangement (Fig.
chronic myelomonocytic leukemia with eosinophilia and an increased 56.19). Serial monitoring with RQ-PCR demonstrates exquisite
incidence (approximately 30%) of T-cell lymphoblastic lymphoma. sensitivity of FIP1L1-PDGFRA–positive patients to low-dose