Page 948 - Hematology_ Basic Principles and Practice ( PDFDrive )
P. 948
Chapter 56 Conventional and Molecular Cytogenomic Basis of Hematologic Malignancies 831
a higher frequency of λ-type MM, high plasma cell-labeling index, and is associated with clonal evolution, drug resistance, and genetic
female predominance, and inferior survival after standard chemo- instability (see Fig. 56.50 fourth row). This deletion is not found in
therapy. Numerous earlier studies have demonstrated that del(13q) patients with other high-risk abnormalities such as t(4;14) or t(14;16),
represents an adverse prognostic marker in patients with MM treated and it appears to be mutually exclusive. Patients with P53 deletion
with conventional chemotherapy. However, more recent studies have have a significantly shorter OS regardless of therapy. The 17p dele-
demonstrated that del(13q) no longer has adverse prognostic signifi- tion, identified by FISH, is considered the most important molecular
cance in patients treated with bortezomib. Molecular cytogenetics cytogenetic factor when determining prognosis.
analyses have revealed that del(13q) is present in 90% of patients with The gain of 1q21 locus has been identified in 45% of cases of
t(4;14) or t(14;16); therefore the apparent adverse impact of del(13q) MGUS, 43% of newly diagnosed MM, and 72% of relapsed MM,
may be related to translocation events. and represents one of the most frequent recurrent chromosomal
Deletion of TP53 at 17p13.1, which occurs in 10% of patients abnormalities in MM (see Fig. 56.50 sixth row and Fig. 56.53). Gain
with MM, is a powerful independent predictor of shortened survival of 1q is observed cytogenetically as an isochromosome, duplications,
del(1p)
del(1p)
A dup 1q B inv dups 1q
der(1;19)(q10;p10)
C der(1;16)(q10;p10) D
1q12~23
der(19) der(21) der(22)
E F
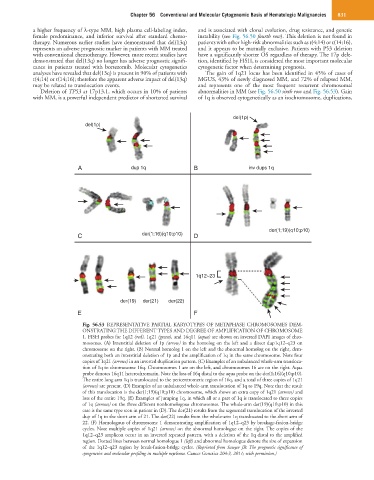
Fig. 56.53 REPRESENTATIVE PARTIAL KARYOTYPES OF METAPHASE CHROMOSOMES DEM-
ONSTRATING THE DIFFERENT TYPES AND DEGREE OF AMPLIFICATION OF CHROMOSOME
1. FISH probes for 1q12 (red), 1q21 (green), and 16q11 (aqua) are shown on inverted DAPI images of chro-
mosomes. (A) Interstitial deletion of 1p (arrow) in the homolog on the left and a direct dup1q12–q23 on
chromosome on the right. (B) Normal homolog 1 on the left and the abnormal homolog on the right, dem-
onstrating both an interstitial deletion of 1p and the amplification of 1q in the same chromosome. Note four
copies of 1q21 (arrows) in an inverted duplication pattern. (C) Examples of an unbalanced whole-arm transloca-
tion of 1q to chromosome 16q. Chromosomes 1 are on the left, and chromosomes 16 are on the right. Aqua
probe denotes 16q11 heterochromatin. Note the loss of 16q distal to the aqua probe on the der(1;16)(q10;p10).
The entire long arm 1q is translocated to the pericentromeric region of 16q, and a total of three copies of 1q21
(arrows) are present. (D) Examples of an unbalanced whole-arm translocation of 1q to 19q. Note that the result
of this translocation is the der(1;19)(q10;p10) chromosome, which shows an extra copy of 1q21 (arrows) and
loss of the entire 19q. (E) Examples of jumping 1q, in which all or a part of 1q is translocated to three copies
of 1q (arrows) on the three different nonhomologous chromosomes. The whole-arm der(19)(q10;p10) in this
case is the same type seen in patient in (D). The der(21) results from the segmental translocation of the inverted
dup of 1q to the short arm of 21. The der(22) results from the whole-arm 1q translocated to the short arm of
22. (F) Homologous of chromosome 1 demonstrating amplification of 1q12–q23 by breakage-fusion-bridge
cycles. Note multiple copies of 1q21 (arrows) on the abnormal homologue on the right. The copies of the
1q12–q23 amplicon occur in an inverted repeated pattern, with a deletion of the 1q distal to the amplified
region. Dotted lines between normal homologue 1 (left) and abnormal homologue denote the size of expansion
of the 1q12–q23 region by break-fusion-bridge cycles. (Reprinted from Sawyer JR: The prognostic significance of
cytogenetics and molecular profiling in multiple myeloma. Cancer Genetics 204:3, 2011; with permission.)