Page 949 - Hematology_ Basic Principles and Practice ( PDFDrive )
P. 949
832 Part VII Hematologic Malignancies
or jumping translocations and is detected with a 1q21-specific FISH
probe. Among 479 patients with newly diagnosed MM, 43% with TCF3-PBX1/1q21
amp lq21 with either a hypodiploid and hyperdiploid karyotype and
del(13q) was associated with poor prognosis as compared with
patients lacking amp1q21 (see Fig. 56.53). In a study of 92 patients
treated with lenalidomide and dexamethasone, del(17p) and gain of
1q21 was associated with a dismal OS. In the most comprehensive
expression profiling survey of patients with MM, reported by the
Arkansas Multiple Myeloma Group using GEP70 (gene expression
profiling of 70 genes), 30% of these genes were located on chromo-
some 1, with most of the downregulated genes located on the short
arms of chromosome 1 and most of the upregulated genes on 1q.
The mechanism for the amplification of 1q is believed to involve
1q12 pericentromeric instability, which most commonly increases the
copy number of 1q by a direct and/or inverted duplication. Further
instability can result in adding a whole-arm segment of 1q to non-
homologous chromosomes by jumping translocations of 1q. The
1q12–q23 amplicon has been reported to be formed by BFB cycles
of the 1q12 pericentromeric heterochromatin and the adjacent bands
of 1q, resulting in an inverted repeat pattern of amplification of the
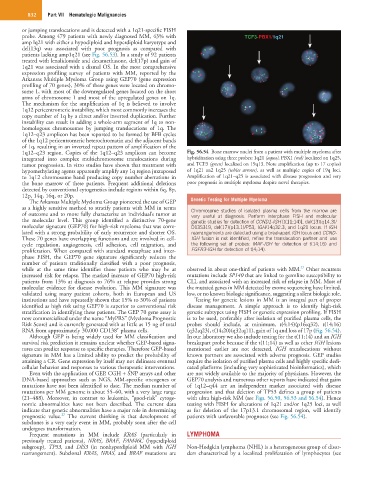
1q12–q23 region. Copies of the 1q12–q23 amplicon can become Fig. 56.54 Bone marrow nuclei from a patient with multiple myeloma after
integrated into complex multichromosome translocations during hybridization using three probes: 1q21 (aqua), PBX1 (red) localized on 1q25,
tumor progression. In vitro studies have shown that treatment with and TCF3 (green) localized on 19q13. Note amplification (up to 17 copies)
hypomethylating agents apparently amplify any 1q region juxtaposed of 1q21 an2 1q25 (white arrows), as well as multiple copies of 19q loci.
to 1q12 chromosome band producing copy number aberrations in Amplification of 1q21–q25 is associated with disease progression and very
the bone marrow of these patients. Frequent additional deletions poor prognosis in multiple myeloma despite novel therapies.
detected by conventional cytogenetics include regions within 6q, 8p,
12p, 14q, 16q, or 20p.
The Arkansas Multiple Myeloma Group pioneered the use of GEP Genetic Testing for Multiple Myeloma
as a highly sensitive method to stratify patients with MM in terms Chromosome studies of isolated plasma cells from the marrow are
of outcome and to more fully characterize an individual’s tumor at very useful at diagnosis. Perform interphase FISH and molecular
the molecular level. This group identified a distinctive 70-gene genetic studies for detection of CCND1-IGH [t(11;14)], del(13)(q14.3)/
molecular signature (GEP70) for high-risk myeloma that was corre- D13S319, del(17)(p13.1)/P53, IGH/14q32.3, and 1q21 locus. If IGH
lated with a strong probability of early recurrence and shorter OS. rearrangements are detected using a breakapart IGH locus and CCND-
These 70 genes have overlapping functions and are involved in cell- IGH fusion is not identified, refine the translocation partner and use
cycle regulation, angiogenesis, cell adhesion, cell migration, and the following set of probes: MAF-IGH for detection of t(14;16) and
proliferation. When compared with standard metaphase and inter- FGFR3-IGH for detection of t(4;14).
phase FISH, the GEP70 gene signature significantly reduces the
number of patients traditionally classified with a poor prognosis,
22
while at the same time identifies those patients who may be at observed in about one-third of patients with MM. Other recurrent
increased risk for relapse. The marked increase of GEP70 high-risk mutations include SP140 that are linked to germline susceptibility to
patients from 13% at diagnosis to 76% at relapse provides strong CLL and associated with an increased risk of relapse in MM. Most of
molecular evidence for disease evolution. This MM signature was the mutated genes in MM detected by exome sequencing have limited,
validated using many patient cohorts, both in European and US low, or no known biologic significance, suggesting a silent biologic role.
institutions and have repeatedly shown that 15% to 30% of patients Testing for genetic lesions in MM is an integral part of proper
identified as high risk using GEP70 is superior to conventional risk disease management. A simple approach is to identify high-risk
stratification in identifying these patients. The GEP 70 gene assay is genetic subtypes using FISH or genetic expression profiling. If FISH
now commercialized under the name “MyPRS” (Myeloma Prognostic is to be used, preferably after isolation of purified plasma cells, the
Risk Score) and is currently generated with as little as 15 ng of total probes should include, at minimum, t(4;14)(p16;q32), t(14;16)
+
RNA from approximately 30,000 CD138 plasma cells. (q32;q23), t(14;20)(q32;q11), gain of 1q and loss of 17p (Fig. 56.54).
Although GEP is being widely used for MM classification and In our laboratory we also include testing for the t(11;14) and an IGH
survival risk prediction it remains unclear whether GEP-based signa- breakapart probe because if the t(11;14) as well as other IGH lesions
tures can predict response to specific therapies. Therefore GEP-based mentioned earlier are not detected, IGH translocations without
signature in MM has a limited ability to predict the probability of known partners are associated with adverse prognosis. GEP studies
attaining a CR. Gene expression by itself may not delineate eventual require the isolation of purified plasma cells and highly specific dedi-
cellular behavior and responses to various therapeutic interventions. cated platforms (including very sophisticated bioinformatics), which
Even with the application of GEP, CGH + SNP arrays and other are not widely available to the majority of physicians. However, the
DNA-based approaches such as NGS, MM-specific oncogenes or GEP70 analysis and numerous other reports have indicated that gains
mutations have not been identified to date. The median number of of 1q12–q44 are an independent marker associated with disease
mutations per MM genome is about 55–60, with a very large range progression and that deletion of TP53 defines a group of patients
(21–488). Moreover, in contrast to leukemia, “good-risk” cytoge- with ultra high-risk MM (see Figs. 56.50, 56.53 and 56.54). Hence
nomic abnormalities have not been described. The current data testing with FISH for alterations of 1q21 and/or 1q25 loci, as well
indicate that genetic abnormalities have a major role in determining as for deletion of the 17p13.1 chromosomal region, will identify
27
prognostic value. The current thinking is that development of patients with unfavorable prognoses (see Fig. 56.54).
subclones is a very early event in MM, probably soon after the cell
undergoes transformation.
Frequent mutations in MM include KRAS (particularly in LYMPHOMA
previously treated patients), NRAS, BRAF, FAM46C (hyperdiploid
subgroup), TP53, and DIS3 (in nonhyperdiploid MM with IGH Non-Hodgkin lymphoma (NHL) is a heterogeneous group of disor-
rearrangement). Subclonal KRAS, NRAS, and BRAF mutations are ders characterized by a localized proliferation of lymphocytes (see