Page 1961 - Williams Hematology ( PDFDrive )
P. 1961
1936 Part XII: Hemostasis and Thrombosis Chapter 113: Molecular Biology and Biochemistry of the Coagulation Factors 1937
In total, 19 SNPs have been identified in the gene encoding TAFI, N-glycosylation sites, with three in the carbohydrate-rich domain
of which six are in the coding region. Of the latter SNPs, two lead to an (Asn96, Asn135, Asn155) and one in the serine protease-binding region
amino acid substitution: an Ala/Thr substitution at position 147 and (Asn192).
a Thr/Ile substitution at position 325. There appears to be a strong
317
correlation between plasma levels of TAFI and polymorphisms in the Antithrombin Function
promoter and 3′-region, but their clinical significance is unclear. The primary proteases targeted by AT are thrombin, factor Xa, and
Epidemiologic studies have indicated that elevated TAFI levels factor IXa. In addition, AT also inhibits factors XIa and XIIa, as well
are correlated with an increased risk of venous thrombosis, albeit that as tissue factor–factor VIIa; however, the latter is only inhibited in the
methods to quantify TAFI or TAFIa have limitations. 318 presence of heparin.
Similar to other serpins, AT acts as a “suicide” substrate for its tar-
INHIBITORS OF COAGULATION: get proteases. These cleave at site Arg393 in the reactive center loop of
AT, upon which AT is, unlike normal substrates, not released, but forms
ANTITHROMBIN, TISSUE FACTOR a 1:1 covalent complex with the protease, thereby blocking the active
PATHWAY INHIBITOR, AND PROTEIN Z/ site. This complex is facilitated by a conformational change of the reac-
tive center loop that folds into the N-terminal region of AT. By doing so,
PROTEIN Z–DEPENDENT PROTEASE the covalently attached protease is dragged along, resulting in distortion
INHIBITOR of its serine protease domain and effectively converting the protease
back into a zymogen-like state.
320
Heparin and related molecules, such as endothelial-bound gly-
ANTITHROMBIN cosaminoglycans, dramatically accelerate the rate of protease inhi-
AT was previously known as AT III as a result of a classification of sev- bition by AT (see Table 113–4) through two distinct mechanisms
eral AT activities in plasma discovered in the 1950s. AT is mainly syn- that characterize inhibition of either factors IXa and Xa or thrombin
319
thesized in the liver and circulates in plasma as a single-chain GP of 432 (Fig. 113–26). In case of the former, binding of a specific pentasaccha-
amino acids (Mr ≈58,000) at a concentration of 2.5 μM with a half-life ride sequence in heparin results in a conformational change in the reac-
of 60 to 70 hours (see Table 113–1). AT is a member of the large serpin tive center loop of AT, which allows for enhanced access by the target
family and is known as SERPINC1 in the systematic nomenclature. protease and is known as allosteric activation of AT (Fig. 113–26, middle
320
panel). This results in a 500-fold acceleration of inhibition of factors
Protein Structure IXa and Xa. The pentasaccharide sequence is present in all forms of
321
AT consists of an N-terminal heparin-binding domain, a carbohy- heparin including low-molecular-weight heparin and fondaparinux, a
drate-rich domain, and a C-terminal serine protease-binding region synthetic pentasaccharide. Heparin-accelerated thrombin inhibition
that comprises the long, flexible, and surface-exposed reactive center involves bridging of AT to the protease, which serves to align the two
loop. Structural stability is provided by three disulfide bonds, two of molecules and enhances the rate of complex formation (Fig. 113–26,
which are located in the N-terminal region and one in the serine pro- right panel). 322,323 This mechanism, also known as the template mech-
tease-binding region. Posttranslational modifications comprise four anism, requires longer heparin molecules found in unfractionated
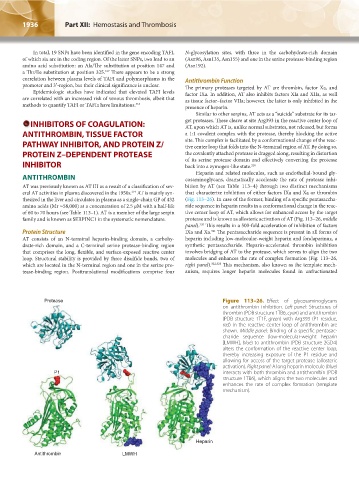
Protease Figure 113–26. Effect of glycosaminoglycans
on antithrombin inhibition. Left panel: Structures of
thrombin (PDB structure 1TB6, cyan) and antithrombin
(PDB structure 1T1F, green) with Arg393 (P1 residue,
red) in the reactive center loop of antithrombin are
shown. Middle panel: Binding of a specific pentasac-
charide sequence (low-molecular-weight heparin
[LMWH], blue) to antithrombin (PDB structure 2GD4)
alters the conformation of the reactive center loop,
thereby increasing exposure of the P1 residue and
allowing for access of the target protease (allosteric
activation). Right panel: A long heparin molecule (blue)
P1 interacts with both thrombin and antithrombin (PDB
structure 1TB6), which aligns the two molecules and
enhances the rate of complex formation (template
mechanism).
Heparin
Antithrombin LMWH
Kaushansky_chapter 113_p1915-1948.indd 1936 9/21/15 2:40 PM