Page 256 - Williams Hematology ( PDFDrive )
P. 256
230 Part IV: Molecular and Cellular Hematology Chapter 16: Cell-Cycle Regulation and Hematologic Disorders 231
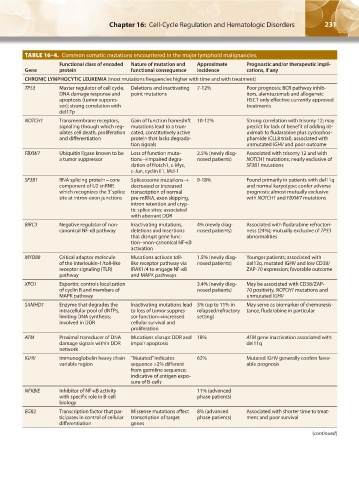
TABLE 16–4. Common somatic mutations encountered in the major lymphoid malignancies.
Functional class of encoded Nature of mutation and Approximate Prognostic and/or therapeutic impli-
Gene protein functional consequence incidence cations, if any
CHRONIC LYMPHOCYTIC LEUKEMIA (most mutations frequencies higher with time and with treatment)
TP53 Master regulator of cell cycle, Deletions and inactivating 7-12% Poor prognosis; BCR pathway inhib-
DNA damage response and point mutations itors, alemtuzumab and allogeneic
apoptosis (tumor suppres- HSCT only effective currently approved
sor); strong correlation with treatments
del17p
NOTCH1 Transmembrane receptors, Gain of function frameshift 10-12% Strong correlation with trisomy 12; may
signaling through which reg- mutations lead to a trun- predict for lack of benefit of adding rit-
ulates cell death, proliferation cated, constitutively active uximab to fludarabine plus cyclophos-
and differentiation protein that lacks degrada- phamide (CLL8 trial); associated with
tion signals unmutated IGHV and poor outcome
FBXW7 Ubiquitin ligase known to be Loss of function muta- 2.5% (newly diag- Associated with trisomy 12 and with
a tumor suppressor tions→impaired degra- nosed patients) NOTCH1 mutations; nearly exclusive of
dation of Notch1, c-Myc, SF3B1 mutations
c-Jun, cyclin E1, Mcl-1
SF3B1 RNA splicing protein – core Spliceosome mutations→ 9-18% Found primarily in patients with del11q
component of U2 snRNP, decreased or increased and normal karyotype; confer adverse
which recognizes the 3’ splice transcription of normal prognosis; almost mutually exclusive
site at intron-exon junctions pre-mRNA, exon skipping, with NOTCH1 and FBXW7 mutations
intron retention and cryp-
tic splice sites; associated
with aberrant DDR
BIRC3 Negative regulator of non- Inactivating mutations, 4% (newly diag- Associated with fludarabine refractori-
canonical NF-κB pathway deletions and insertions nosed patients) ness (24%); mutually exclusive of TP53
that disrupt gene func- abnormalities
tion→non-canonical NF-κB
activation
MYD88 Critical adaptor molecule Mutations activate toll- 1.5% (newly diag- Younger patients; associated with
of the interleukin-1/toll-like like receptor pathway via nosed patients) del13q, mutated IGHV and low CD38/
receptor signaling (TLR) IRAK1/4 to engage NF-κB ZAP-70 expression; favorable outcome
pathway and MAPK pathways
XPO1 Exportin; controls localization 3.4% (newly diag- May be associated with CD38/ZAP-
of cyclin B and members of nosed patients) 70 positivity, NOTCH1 mutations and
MAPK pathway unmutated IGHV
SAMHD1 Enzyme that degrades the Inactivating mutations lead 3% (up to 11% in May serve as biomarker of chemoresis-
intracellular pool of dNTPs, to loss of tumor suppres- relapsed/refractory tance, fludarabine in particular
limiting DNA synthesis; sor function→increased setting)
involved in DDR cellular survival and
proliferation
ATM Proximal transducer of DNA Mutations disrupt DDR and 18% ATM gene inactivation associated with
damage signals within DDR impair apoptosis del11q
network
IGHV Immunoglobulin heavy chain “Mutated” indicates 62% Mutated IGHV generally confers favor-
variable region sequence >2% different able prognosis
from germline sequence;
indicative of antigen expo-
sure of B-cells
NFKBIE Inhibitor of NF-κB activity 11% (advanced
with specific role in B-cell phase patients)
biology
EGR2 Transcription factor that par- Missense mutations affect 8% (advanced Associated with shorter time to treat-
ticipates in control of cellular transcription of target phase patients) ment and poor survival
differentiation genes
(continued)
Kaushansky_chapter 16_p0213-0246.indd 231 9/18/15 11:57 PM