Page 912 - Hematology_ Basic Principles and Practice ( PDFDrive )
P. 912
Chapter 56 Conventional and Molecular Cytogenomic Basis of Hematologic Malignancies 795
disease (p = .0002) but comparable to the poor-risk group (p = .97). at approximately 70.9 Mb (five cases) and 72.1 Mb (seven cases) on
Trisomy 11 is associated with clinical aggressiveness and represents a the X chromosome. None of the 11 breakpoints occurred in a gene,
high-risk cytogenetic abnormality. strongly indicating that the idic(X)(q13) does not result in a fusion
Deletions of 17p are seen in 3% to 4% of MDS and AML gene. Instead, the functional outcome of the abnormality confers a
patients. These patients often display several other chromosomal gene dosage effect because of the concurrent gain of Xpter-q13 and
rearrangements, including monosomy 17, isochromosome 17q (see loss of Xq13-qter. This region of the X chromosome is enriched for
Fig. 56.20, third row), and unbalanced translocations between chro- repeated sequences and most likely, these repeats may facilitate the
mosome 17 and another chromosome. Approximately 30% of these formation of idic(X). The isodicentric X chromosome was inactive in
deletions are related to therapy. The extent of 17p deletion in all cases some patients and active in other patients; hence idic(X) appears to
involves the TP53 gene. There appears to be a close correlation be leukemogenic regardless of X a or X i involvement.
between dysgranulopoiesis (e.g., pseudo–Pelger-Huet hypolobula- Gain of 1q, usually in the form of an unbalanced translocation,
tion) and small vacuoles in neutrophils with 17p abnormalities and is a recurrent abnormality in MDS and appears to be a marker of
TP53 deletion. The median survival of these patients is poor. disease progression. Specifically, the gain of 1q in the form of jumping
An isodicentric X chromosome in Xq13-idic(X) is a rare but translocations either at diagnosis or the subsequent acquisition of
recurrent abnormality that has been associated with refractory anemia jumping 1q translocations appears to be associated with imminent
with ringed sideroblasts. All MDS patients with idic(X)(q13) are transformation to AML in patients after an average of 9 months (Fig.
females, most likely because the formation of idic(X) would result in 56.22). Once acquired, the patient’s prognosis tended to be depen-
nullisomy for Xq13-qter in males. The median age at the time of dent on the copy number of 1q, indicating that gain of jumping 1q
diagnosis is 73.5 years. The outcome of idic(X)-positive cases is translocations was associated with both disease progression and poor
variable; some investigators report aggressive and rapidly fatal disease prognosis. In MDS, the average time to develop jumping 1q was less
and others a relatively favorable clinical course with survival for than 2 years. Treatment (azacitidine, chemotherapy, or stem cell
several years. The fact that idic(X) most often occurs as the sole transplant) may temporarily reduce or eradicate the clone, but the
cytogenetic abnormality suggests that it may in itself be sufficient for responses were not durable. These findings did not provide any
leukemogenesis. Using a high-resolution SNP array, the breakpoints common pattern as to the role of the partner chromosomes in unbal-
on idic(X)(q13) were mapped into two distinct breakpoint clusters, anced jumping 1q translocations. However, 81% of recipient
D
A dup(1)(q12q21)
der(13)t(1;3)(q12;q34)
E
der(9)(t(1;9)(q12;q12)
B
der(6)t(1;6)(q21;p23)
C F
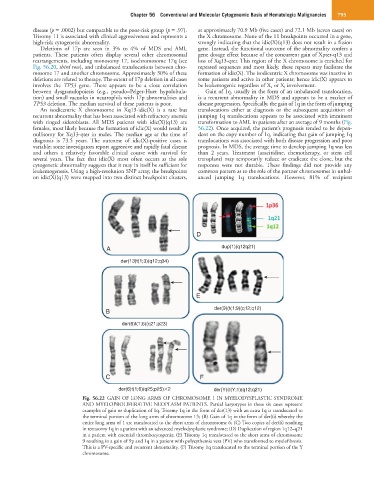
der(6)t(1;6)(q25;p25) 2 der(Y)(t(Y;1)(q12;q21)
Fig. 56.22 GAIN OF LONG ARMS OF CHROMOSOME 1 IN MYELODYSPLASTIC SYNDROME
AND MYELOPROLIFERATIVE NEOPLASM PATIENTS. Partial karyotypes in these six cases represent
examples of gain or duplication of 1q. Trisomy 1q in the form of der(13) with an extra 1q is translocated to
the terminal portion of the long arms of chromosome 13; (B) Gain of 1q in the form of der(6) whereby the
entire long arms of 1 are translocated to the short arms of chromosome 6; (C) Two copies of der(6) resulting
in tetrasomy 1q in a patient with an advanced myelodysplastic syndrome; (D) Duplication of region 1q12–q21
in a patient with essential thrombocytopenia; (E) Trisomy 1q translocated to the short arms of chromosome
9 resulting in a gain of 9p and 1q in a patient with polycythemia vera (PV) who transformed to myelofibrosis.
This is a PV-specific and recurrent abnormality. (F) Trisomy 1q translocated to the terminal portion of the Y
chromosome.